

Promega Restriction Enzyme Tool. You can find an enzyme by entering the name, recognition sequence or overhang sequence.
Promega enzymes are indicated in orange. Search by Name Select "Name", and enter the name of your enzyme into the searchbox. Search by Sequence Select "Sequence", and enter the recognition sequence into the searchbox. N = A,T,C or G (any nucleotide) Y = T or C (pyrimidine) R = G or A (purine) M = A or C (amino) K = G or T (keto) S = G or C (strong interactions: 3 H bonds) W = A or T (weak interactions: 2 H bonds) B = G, T or C (not A) V = G, C or A (not T or U) D = G, A or T (not C) H = A, C or T (not G) Search by Overhang Search by specific 3' or 5' overhang, or list all blunt enzymes.
Find Buffers For Double Digests Once you have selected an enzyme, you can select a second enzyme and retrieve information on suitable buffers for double digests. Metabolite fingerprinting of pennycress (Thlaspi arvense L.) embryos to assess active pathways during oil synthesis. + Author Affiliations ↵* To whom correspondence should be addressed.

E-mail: alonso.19@osu.edu Abstract Pennycress (Thlaspi arvense L.), a plant naturalized to North America, accumulates high levels of erucic acid in its seeds, which makes it a promising biodiesel and industrial crop. The main carbon sinks in pennycress embryos were found to be proteins, fatty acids, and cell wall, which respectively represented 38.5, 33.2, and 27.0% of the biomass at 21 days after pollination.
Key words: Introduction Petroleum is the largest energy source in the USA, accounting for 28% of all energy consumed in 2013 (www.eia.gov). Field pennycress (Thlaspi arvense L.; Supplementary Fig. In plants, different pathways in central metabolism are responsible for allocating the carbon skeletons, reducing power and energy required for fatty acid synthesis. Materials and methods Chemicals Metabolite standards, 3 N methanolic HCl, and toluene were purchased from Sigma. Plant growth Biomass extraction Oil Derivatization. Optimal Percent Agarose Concentration.
Customer Log In.
Logga in på Office 365. Ange e-postadressen för det konto som du vill logga in med.
Vi kan inte hitta ditt konto. Vilken typ av konto vill du använda? Centrifuge Rotor Speed Calculator. Simple Online Collaboration: Online File Storage, FTP Replacement, Team Workspaces. European hazard symbols. The Hazard symbols for chemicals are pictograms defined by the European community for labeling chemical packagings (for storage and workplace) and containers (for transportation).
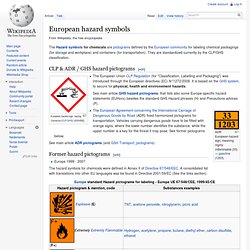
They are standardized currently by the CLP/GHS classification. CLP & ADR / GHS hazard pictograms[edit] European hazard sign, saying Corrosive (CLP/GHS) [GSH05]) The European Union CLP Regulation (for "Classification, Labelling and Packaging") was introduced through the European directives (EC) N°1272/2008. It is based on the GHS system, to secure for physical, health and environement hazards. See main article GHS hazard pictograms, that lists also some Europe-specific hazard statements (EUHxxx) besides the standard GHS Hazard phrases (H) and Precautions advices (P). Outlook Web App. Chemical Products Search.
ORFs, Splicing & Coding. ORFs, Start Sites & Coding Regions The ORF Finder (Open Reading Frame Finder) is a graphical analysis tool which finds all open reading frames of a selectable minimum size in a user's sequence or in a sequence already in the database.

This server provides access to the program Genscan for predicting the locations and exon-intron structures of genes in genomic sequences from a variety of organisms. This server can accept sequences up to 1 million base pairs (1 Mbp) in length. The NetStart server produces neural network predictions of translation start in vertebrate and Arabidopsis thaliana nucleotide sequences. Projector is a program for the comparative, homology based prediction of protein coding genes in mouse and human DNA.
Online ORF analysis giving detailed report. HMM-based gene structure prediction (multiple genes, both chains) Protea is a software devoted to protein-coding sequences identification. Gene identification in novel eukaryotic genomes by self-training algorithm. Get_simple.
Bibliographie. Bioinformatique. INRA Versailles.