

DNA sequencing. DNA sequencing is the process of determining the precise order of nucleotides within a DNA molecule.
It includes any method or technology that is used to determine the order of the four bases—adenine, guanine, cytosine, and thymine—in a strand of DNA. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery. Knowledge of DNA sequences has become indispensable for basic biological research, and in numerous applied fields such as diagnostic, biotechnology, forensic biology, and biological systematics. The rapid speed of sequencing attained with modern DNA sequencing technology has been instrumental in the sequencing of complete DNA sequences, or genomes of numerous types and species of life, including the human genome and other complete DNA sequences of many animal, plant, and microbial species.
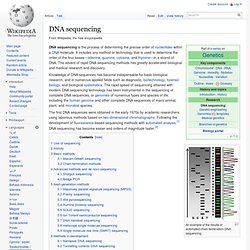
An example of the results of automated chain-termination DNA sequencing. Use of sequencing[edit] History[edit] Basic methods[edit] One cell is all you need. The notion that police can identify a suspect based on the tiniest drop of blood or trace of tissue has long been a staple of TV dramas, but scientists at Harvard have taken the idea a step further.

Using just a single human cell, they can reproduce an individual’s entire genome. As described in a Dec. 21 paper in Science, a team of researchers, led by Xiaoliang Sunney Xie, the Mallinckrodt Professor of Chemistry and Chemical Biology, and made up of postdoctoral fellow Chenghang Zong, graduate student Alec Chapman, and former graduate student Sijia Lu, developed a method — dubbed MALBAC, short for Multiple Annealing and Looping-based Amplification Cycles — that requires just one cell to reproduce an entire DNA molecule. More than three years in the making, the breakthrough technique offers the potential for early cancer treatment by allowing doctors to obtain a genetic “fingerprint” of a person’s cancer from circulating tumor cells. Perfection Is Skin Deep: Everyone Has Flawed Genes : Shots - Health News. Hide captionWhen researchers looked at the genetic sequences of 179 individuals, they found far more defects in the patterns of As, Ts, Gs, and Cs than they expected. iStockphoto.com When researchers looked at the genetic sequences of 179 individuals, they found far more defects in the patterns of As, Ts, Gs, and Cs than they expected.
We all know that nobody's perfect. But now scientists have documented that fact on a genetic level. Researchers discovered that normal, healthy people are walking around with a surprisingly large number of mutations in their genes. It's been well known that everyone has flaws in their DNA, though, for the most part, the defects are harmless. "It's such an interesting question that people had been trying to make estimates from indirect approaches for a long time," says Chris Tyler-Smith of the Wellcome Trust Sanger Institute in Cambridge, England.
But Tyler-Smith and his colleagues wanted to get a more precise, direct estimate. "We're all mutants. " Genomic data are growing, but what do we really know? "We live in the post-genomic era, when DNA sequence data is growing exponentially", says Miami University (Ohio) computational biologist Iddo Friedberg.
"But for most of the genes that we identify, we have no idea of their biological functions. They are like words in a foreign language, waiting to be deciphered. " Understanding the function of genes is a problem that has emerged at the forefront of molecular biology. Many groups develop and employ sophisticated algorithms to decipher these "words". However, until now there was no comprehensive picture of how well these methods perform, "To use the information in our genes to our advantage, we first need to take stock of how well we are doing in interpreting these data". Bioinformatics and computational biology : Charting a course for genomic medicine from base pairs to bedside : Nature. The major bottleneck in genome sequencing is no longer data generation—the computational challenges around data analysis, display and integration are now rate limiting.

New approaches and methods are required to meet these challenges. Data analysis. Computational tools are quickly becoming inadequate for analysing the amount of genomic data that can now be generated, and this mismatch will worsen. Innovative approaches to analysis, involving close coupling with data production, are essential. Data integration. Visualization. Computational tools and infrastructure. Training.