

The ConSurf Server. The SELECTON Server. RAxML BlackBox. ATGC: PhyML. IDT - PrimerQuest Input. Primer3 Input (version 0.4.0) Copyright Notice and Disclaimer Copyright (c) 1996,1997,1998,1999,2000,2001,2004,2006,2007 Whitehead Institute for Biomedical Research, Steve Rozen, Maido Remm, Triinu Koressaar and Helen Skaletsky All rights reserved.
Redistribution and use in source and binary forms, with or without modification, are permitted provided that the following conditions are met: * Redistributions of source code must retain the above copyright notice, this list of conditions and the following disclaimer. * Redistributions in binary form must reproduce the above copyright notice, this list of conditions and the following disclaimer in the documentation and/or other materials provided with the distribution. * Neither the names of the copyright holders nor contributors may be used to endorse or promote products derived from this software without specific prior written permission.
Citing Primer3 We request that use of this software be cited in publications as Source code available at primer3.sourceforge.net/. Saccharomyces Genome Database. Tree of Life Web Project. The Tree of Life Web Project (ToL) is a collaborative effort of biologists and nature enthusiasts from around the world.
On more than 10,000 World Wide Web pages, the project provides information about biodiversity, the characteristics of different groups of organisms, and their evolutionary history (phylogeny). Each page contains information about a particular group, e.g., salamanders, segmented worms, phlox flowers, tyrannosaurs, euglenids, Heliconius butterflies, club fungi, or the vampire squid. ToL pages are linked one to another hierarchically, in the form of the evolutionary tree of life. Starting with the root of all Life on Earth and moving out along diverging branches to individual species, the structure of the ToL project thus illustrates the genetic connections between all living things.
Trex-online. Home - Taxonomy - NCBI. KEGG PATHWAY Database. STRING: functional protein association networks. DAVID Functional Annotation Bioinformatics Microarray Analysis. Charting Pathways of Life. UniProtKB/TrEMBL Release Statistics. UniProtKB/TrEMBL PROTEIN DATABASE RELEASE 2014_04 STATISTICS 1.
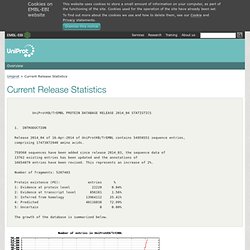
INTRODUCTION Release 2014_04 of 16-Apr-2014 of UniProtKB/TrEMBL contains 54958551 sequence entries, comprising 17473872940 amino acids. 759560 sequences have been added since release 2014_03, the sequence data of 13762 existing entries has been updated and the annotations of 16654879 entries have been revised. This represents an increase of 2%. Number of fragments: 5207483 Protein existence (PE): entries % 1: Evidence at protein level 22220 0.04% 2: Evidence at transcript level 856181 1.56% 3: Inferred from homology 13964112 25.41% 4: Predicted 40116038 72.99% 5: Uncertain 0 0.00% The growth of the database is summarized below. Www.pymol.org. UniProt. MUSCLE Multiple Accurate and Fast Sequence Comparison by Log-Expectation Tool. DaliLite Pairwise Alignment Tool for Protein Structures.
Protein Data Bank - RCSB PDB. A Structural View of Biology This resource is powered by the Protein Data Bank archive-information about the 3D shapes of proteins, nucleic acids, and complex assemblies that helps students and researchers understand all aspects of biomedicine and agriculture, from protein synthesis to health and disease.
As a member of the wwPDB, the RCSB PDB curates and annotates PDB data. The RCSB PDB builds upon the data by creating tools and resources for research and education in molecular biology, structural biology, computational biology, and beyond. Pfam: Home page. TMHMM Server, v. 2.0. SWISS-MODEL Workspace. GetArea. ProtScale. PredictProtein - Sequence Analysis, Structure and Function Prediction. Nucleic Acids Research. GenBank Home. RefSeq: NCBI Reference Sequence Database. BLAST: Basic Local Alignment Search Tool. Bioinformatics Tools for Pairwise Sequence Alignment. Pairwise Sequence Alignment is used to identify regions of similarity that may indicate functional, structural and/or evolutionary relationships between two biological sequences (protein or nucleic acid). By contrast, Multiple Sequence Alignment (MSA) is the alignment of three or more biological sequences of similar length. From the output of MSA applications, homology can be inferred and the evolutionary relationship between the sequences studied.
Global Alignment Global alignment tools create an end-to-end alignment of the sequences to be aligned. There are separate forms for protein or nucleotide sequences. Needle EMBOSS Needle creates an optimal global alignment of two sequences using the Needleman-Wunsch algorithm. Goldman Group Software- webPRANK. For feedback, questions and comments, contact Ari Loytynoja at ari@ebi.ac.uk.
The alignment program PRANK and its graphical version PRANKSTER are available as stand-alone programs for different operating systems. Find more informations at the program home pages. The PRANK alignment algorithm is described in these papers. Our thanks go to the EBI External Services team, who provide the infrastucture, help and support upon which the PRANK web server relies heavily. However, they are not responsible for any problems that may occur and should not be addressed with complaints or requests concerning this service. Align Sequences using ClustalW2.
Goldman Group Software- webPRANK. BioEdit Sequence Alignment Editor for Windows 95/98/NT/XP/Vista/7. Are you having trouble with the Right-click to change the reference sequence for the dot-conservation view because of the context menu feature?

You can now use Ctrl+Right Click to change the dot-conservation referene sequence. Notes for 64-bit OS users: For Windows 7 or 8, 64-bit users of readSeq (automated conversion from file formats such as nexus for import or export into and out of BioEdit), the version of readSeq originally created by Don Gilbert that is packaged with BioEdit will not run in 64-bit Windows. A 64-bit-compatible port has been created by Mathew D. Pagel and is packaged with BioEdit in its original distribution package "ReadSeqv202.zip" with a small block of text added to the top of the original "Instructions.txt" file included in the zip file distribution.
MAFFT alignment and NJ / UPGMA phylogeny. Input: Paste protein or DNA sequences in fasta format.
Example or upload a plain text file: Structural alignment 1 (optional): Paste an alignment in fasta format. ExampleThese sequences will be aligned with the 'input' sequences above, being used as a constraint. Genome Browser. Restriction Maps. National Center for Biotechnology Information. Google Scholar. Free Bioinformatics Educational Resource. OMIM Home.