

Evolutionary loss of melanogenesis in the tunicate Molgula occulta. The CNS connectome of a tadpole larva of Ciona intestinalis (L.) highlights sidedness in the brain of a chordate sibling. Reviewer #1: Ryan et al. present the result of a tremendous effort reconstructing the connectome of larval Ciona intestinalis from a series of electron micrographs.
This represents the second whole-organism connectome ever fully reconstructed and is a finding of major significance. The authors relate their findings within the context of structural and connectivity asymmetries, particularly emphasizing relevant comparisons to vertebrates. As many readers are unlikely to be familiar with C. intestinalis, the Introduction should discuss the life cycle of the larva and its behavior. My understanding is that younger larvae swim upwards and older larvae swim downwards to eventually settle based in part on sensors for gravity and light. We now added the following text to the Introduction: In addition, we address the issue of how the Ciona connectome relates to its behaviors, later, in the Discussion. Some relevant comparisons to other connectomics work are missing. Reviewer #2:
Amyloid beta. Amyloid beta (Aβ or Abeta) denotes peptides of 36–43 amino acids that are crucially involved in Alzheimer's disease as the main component of the amyloid plaques found in the brains of Alzheimer patients.[2] The peptides derive from the amyloid precursor protein (APP), which is cleaved by beta secretase and gamma secretase to yield Aβ.
Aβ molecules can aggregate to form flexible soluble oligomers which may exist in several forms. It is now believed that certain misfolded oligomers (known as "seeds") can induce other Aβ molecules to also take the misfolded oligomeric form, leading to a chain reaction akin to a prion infection. The oligomers are toxic to nerve cells.[3] The other protein implicated in Alzheimer's disease, tau protein, also forms such prion-like misfolded oligomers, and there is some evidence that misfolded Aβ can induce tau to misfold.[4][5] A recent study suggested that APP and its amyloid potential is of ancient origins, dating as far back as early deuterostomes.[6] Amyloid precursor protein.
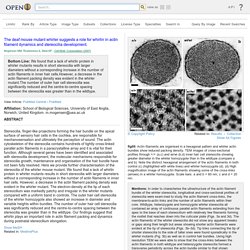
(a) A low magnification image immediately after co-injection of red negatively charged and green glycine-conjugated beads showing the injection site, marked with an oil droplet, appearing as a round yellow sphere.
Overlap of red and green fluorescence produces a yellow image. (b) At 50 min after injection, the red carboxylated beads have progressed in the anterograde direction (to the right) while the green glycine-conjugated beads have made no progress. (c)–(e) An axon co-injected with red APP-C beads and green glycine beads and imaged for 100 frames at 4 s intervals at 40× magnification.
(c) Red channel (left) first frame; (center) 50 frames superimposed; (right) all 100 frames superimposed. Note the progression of individual beads towards the right, anterograde, side of the injection site heading towards the presynaptic terminal. Actin filaments are organised in a hexagonal pattern an. The deaf mouse mutant whirler suggests a role for whirlin in actin filament dynamics and stereocilia development.
Mogensen MM, Rzadzinska A, Steel KP - Cell Motil. Cytoskeleton (2007) Bottom Line: We found that a lack of whirlin protein in whirler mutants results in short stereocilia with larger diameters without a corresponding increase in the number of actin filaments in inner hair cells.However, a decrease in the actin filament packing density was evident in the whirler mutant.The number of outer hair cell stereocilia was significantly reduced and the centre-to-centre spacing between the stereocilia was greater than in the wildtype. Affiliation: School of Biological Sciences, University of East Anglia, Norwich, United Kingdom. m.mogensen@uea.ac.uk. Kinocilium. A kinocilium is a special type of cilium on the apex of hair cells located in the sensory epithelium of the vertebrate inner ear.
Evolution of flagella. The evolution of flagella is of great interest to biologists because the three known varieties of flagella (eukaryotic, bacterial, and archaeal) each represent a sophisticated cellular structure that requires the interaction of many different systems.
Eukaryotic flagellum[edit] There are two competing groups of models for the evolutionary origin of the eukaryotic flagellum (referred to as cilium below to distinguish it from its bacterial counterpart). Recent studies on the microtubule organizing center (MTOC) suggest that the most recent ancestor of all eukaryotes already had a complex flagellar apparatus.[1] Endogenous, autogenous and direct filiation models[edit] Prokaryotic cytoskeleton. Elements of the Caulobacter crescentus cytoskeleton.
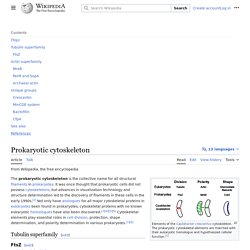
The prokaryotic cytoskeletal elements are matched with their eukaryotic homologue and hypothesized cellular function.[1] Symbiogenesis. Symbiogenesis is the merging of two separate organisms to form a single new organism.

The idea originated with Konstantin Mereschkowsky in his 1926 book Symbiogenesis and the Origin of Species, which proposed that chloroplasts originate from cyanobacteria captured by a protozoan.[1] Ivan Wallin also supported this concept in his book "Symbionticism and the Origins of Species". He suggested that bacteria might be the cause of the origin of species, and that species creation may occur through endosymbiosis. Today both chloroplasts and mitochondria are believed, by those who ascribe to the endosymbiotic theory, to have such an origin. A fundamental principle of modern evolutionary theory is that mutations arise one at a time and either spread through the population or not, depending on whether they offer an individual fitness advantage. Undulipodium. Diagram of a cross-section of the axoneme microtubule array present in all undulipodia. 1-A. and 1-B.
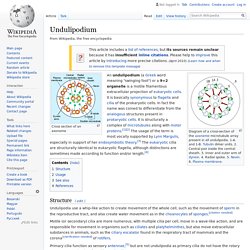
Tubulin dimer units. 2. Central pair inside the central sheath. 3. Inner and outer arm of dynein. 4. Radial spoke. 5. Cilium. There are two types of cilia: motile cilia and nonmotile, or primary, cilia, which typically serve as sensory organelles.
In eukaryotes, motile cilia and flagella together make up a group of organelles known as undulipodia.[4] Eukaryotic cilia are structurally identical to eukaryotic flagella, although distinctions are sometimes made according to function and/or length.[5] Biologists have various ideas about how the various flagella may have evolved. Types and distribution[edit] Cilia can be divided into primary forms and motile forms.[6] Primary/Immotile cilia[edit] In animals, primary cilia are found on nearly every cell.[2] In comparison to motile cilia, non-motile (or primary) cilia usually occur one per cell; nearly all mammalian cells have a single non-motile primary cilium.
Illustration depicting motile cilia. Evolution of flagella. Timeline of the formation of the Universe. From Wikipedia, the free encyclopedia. E2F. E2F family[edit] Schematic diagram of the amino acid sequences of E2F family members (N-terminus to the left, C-terminus to the right) highlighting the relative locations of functional domains within each member: Genes[edit] Homo sapiens E2F1 mRNA or E2F1 protein sequences from NCBI protein and nucleotide database. PAX7. Paired box protein Pax-7 is a protein that in humans is encoded by the PAX7 gene.[5][6][7] Function[edit] Pax-7 plays a role in neural crest development and gastrulation, and it is an important factor in the expression of neural crest markers such as Slug, Sox9, Sox10 and HNK-1.[8] PAX7 is expressed in the palatal shelf of the maxilla, Meckel's cartilage, mesencephalon, nasal cavity, nasal epithelium, nasal capsule and pons. PAX6. Paired box protein Pax-6, also known as aniridia type II protein (AN2) or oculorhombin, is a protein that in humans is encoded by the PAX6 gene.[5] Pax6 is a transcription factor present during embryonic development.
The encoded protein contains two different binding sites that are known to bind DNA and function as regulators of gene transcription. It is a key regulatory gene of eye and brain development. Within the brain, the protein is involved in development of the specialized cells that process smell. As a transcription factor, Pax6 activates and/or deactivates gene expression patterns to ensure for proper development of the tissue. GetSharedSiteSession?rc=1&redirect= Identification of cVA Opsin Additional bioinformatic analyses of other genome databases allowed us to identify VA-like genes in multiple vertebrate classes, including the Amphibia (Xenopus laevis), the Reptilia (Anolis carolinensis), and the Elasmobranchii (Callorhinchus milii), but failed to find any VA homologs within the mammalian databases.
A phylogenetic analysis of the deduced VA proteins places the newly identified sequences in a separate clade (Figure 1Figure 1). Figure 1 Phylogenetic Analysis of VA Opsins. International Journal of Biological Sciences 02: 0038 image No. 02. Gap junction - Wikipedia. One gap junction channel is composed of two connexons (or hemichannels), which connect across the intercellular space.[4][5][6] Gap junctions are analogous to the plasmodesmata that join plant cells.[7] Gap junctions occur in virtually all tissues of the body, with the exception of adult fully developed skeletal muscle and mobile cell types such as sperm or erythrocytes. Gap junctions, however, are not found in simpler organisms such as sponges and slime molds.
A gap junction may also be called a nexus or macula communicans. While an ephapse has some similarities to a gap junction, by modern definition the two are different. Category Results. LARVAE OF PORIFERA-COELENTERATA-PLATY HELMENTHES. Epiblast - Embryonic Development & Stem Cells - LifeMap Discovery. Dopamine. We ask you, humbly, to help. Hi reader in Canada, it seems you use Wikipedia a lot; that's great! It's a little awkward to ask, but this Wednesday we need your help. We’re not salespeople. We’re librarians, archivists, and information junkies. Melanin - Wikipedia. Melatonin. Enterochromaffin cell - Wikipedia. Enterochromaffin (EC) cells (also known as Kulchitsky cells), discovered by Nikolai Kulchitsky of Karazin Kharkiv National University.[1] They are a type of enteroendocrine and neuroendocrine cell. Serotonin - Wikipedia. Monoamine neurotransmitter Serotonin ([6][7][8]) or 5-hydroxytryptamine (5-HT) is a monoamine neurotransmitter.
It has a popular image as a contributor to feelings of well-being and happiness, though its actual biological function is complex and multifaceted, modulating cognition, reward, learning, memory, and numerous physiological processes such as vomiting and vasoconstriction.[9] Tunicate ampullae. TUNICATE CHIMERAS. L-DOPA - Wikipedia. Medical use[edit] Neuromelanin - Wikipedia. L-DOPA - Wikipedia. Five Kingdom Classification System. Chapter 12A. Plant Development. The Structure and Functions of Flowers. Developmental Biology 3230. Introductionto bilateria 2012. GEOL 331 Principles of Paleontology. Developmental Biology in the Ocean 2013: Nematostella vectensis. Nematostella - Wikipedia. Bilateria. Nematostella. Volvox - Wikipedia. Volvox - Volvox - Wikipedia. WVGES Mini-Museum, What's New?
The Artful Amoeba - Scientific American Blog Network. Theory of evolution. Learn Science at Scitable. GlycoWord / Evolution-ES-A02. Choanoflagellates. PubMed Central, Fig. 2: Cell Mol Life Sci. 2011 Oct; 68(19): 3201–3207. Published online 2011 Aug 11. doi: 10.1007/s00018-011-0784-5. WoRMS - World Register of Marine Species - Synoicum adareanum (Herdman, 1902) Fossil Octopuses. Squid Evolution - Squid Facts and Information. iBiology: Bringing the world's best biology videos to you. Science Education - National Institute of General Medical Sciences. Neoteny. Atlas of Human Embryos [by: RF Gasser, PhD.] - Ch.5. What can we learn about our limbs from the limbless? Lines of Evidence: Developmental Biology, Page 1 of 2. Unicellular Organisms – The Cambrian Explosion – Nano Machines. Chlorocruorin. Red Blood Cells. Neanderthal and Denisovan Genomes.
Aragonite sea. Supercontinent cycle. Pangaea. October 12, 2009, 364 (1531)