

Using R Markdown – RStudio Support. R Markdown enables easy authoring of reproducible web reports from R.
It offers: Easy creation of web reports from R that can be automatically regenerated whenever underlying code or data changes. A highly accessible syntax (markdown) which lower the barriers to entry for reproducible research. Output of a standalone HTML file (with images embedded directly in the file) that is easy to share using email, Dropbox, or by deploying to a web server. Support for publishing dynamic and interactive web content. Embedding Markdown in Jekyll HTML - Stack Overflow. Document Templates. Overview If there is a particular form of R Markdown document that you or those you work with create frequently, it may make sense to create a re-usable document template for it.
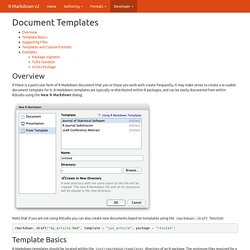
R Markdown templates are typically re-distributed within R packages, and can be easily discovered from within RStudio using the New R Markdown dialog: Note that if you are not using RStudio you can also create new documents based on templates using the rmarkdown::draft function: rmarkdown::draft("my_article.Rmd", template = "jss_article", package = "rticles") Template Basics. Jimhester/knitrBootstrap · GitHub. Pandoc - Pandoc User’s Guide. Pandoc [options] [input-file]… Pandoc is a Haskell library for converting from one markup format to another, and a command-line tool that uses this library.

It can read markdown and (subsets of) Textile, reStructuredText, HTML, LaTeX, MediaWiki markup, Haddock markup, OPML, Emacs Org-mode and DocBook; and it can write plain text, markdown, reStructuredText, XHTML, HTML 5, LaTeX (including beamer slide shows), ConTeXt, RTF, OPML, DocBook, OpenDocument, ODT, Word docx, GNU Texinfo, MediaWiki markup, EPUB (v2 or v3), FictionBook2, Textile, groff man pages, Emacs Org-Mode, AsciiDoc, InDesign ICML, and Slidy, Slideous, DZSlides, reveal.js or S5 HTML slide shows.
It can also produce PDF output on systems where LaTeX is installed. Pander: A Pandoc writer in R. The main aim of the pander R package is to provide a minimal and easy tool for rendering R objects into Pandoc's markdown.

The package is also capable of exporting/converting complex Pandoc documents (reports) in various ways. Regarding the difference between pander and other packages for exporting R objects to different file formats, please refer to this section. Current build and test coverage status: Some CRAN statistics: The stable version. Rmarkdown: Alter Action Depending on Document. Can I see a show of hands for those who love rmarkdown?
Yeah me too. One nifty feature is the ability to specify various document prettifications in the YAML of a .Rmd document and then use: The Problem. Lot of reports with a single click! Suppose you want to create a huge number of pdf files through RMarkdown and pandoc, each of them including a statistical analysis on a part of your data, for example on each row of your data frame.
You need to write a .R file with cycles from 1 to the number of rows of your data set the instructions contained in a .Rmd file. Suppose that: your data set name is data.csv and its path is datapath/data.csv’’your .Rmd file name is report.Rmd and its path is basepath/report.Rmd’’ One weird trick to compile multipartite dynamic... < biochemistries. This afternoon I stumbled across this one weird trick an undocumented part of the YAML headers that get processed when you click the ‘knit’ button in RStudio.
Knitting turns an Rmarkdown document into a specified format, using the rmarkdown package’s render function to call pandoc (a universal document converter written in Haskell). If you specify a knit: field in an Rmarkdown YAML header you can replace the default function (rmarkdown::render) that the input file and encoding are passed to with any arbitrarily complex function. For example, the developer of slidify passed in a totally different function rather than render - slidify::knit2slides. I thought it’d be worthwhile to modify what was triggered upon clicking that button - as simply as using a specified output file name (see StackOverflow here), or essentially running a sort of make to compose a multi-partite document. via Software Carpentry.
Makefiles and RMarkdown. Quite some time ago (October 2013, according to Amazon), I bought a copy of “Reproducible Research with R and RStudio” by Christopher Gandrud.
And it was awesome. Since then, I’ve been using knitr and RMarkdown quite a lot. However, until recently, I never bothered with a makefile. At the time, I had assumed that it was something only available to people on *nix systems and back then I was developing exclusively on PC. I even wrote some R scripts that were more or less makefiles; reading the contents of a directory, checking for output and running render or knit or whatever.
So, enough preamble. The benefit A makefile ensures that all of your work gets done, but only when it needs to and that each step has the raw material it needs to work. The assumptions Use RMarkdown files as your default. The steps. Make prettier documents by reusing chunks in RMarkdown. No revelations here, just a little R tip for generating more readable documents.
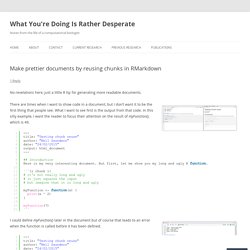
Original with lots of code at the top There are times when I want to show code in a document, but I don’t want it to be the first thing that people see. What I want to see first is the output from that code. In this silly example, I want the reader to focus their attention on the result of myFunction(), which is 49. Function use before definition = error I could define myFunction() later in the document but of course that leads to an error when the function is called before it has been defined. Solution: use the chunk option ref.label to call chunk 2 from chunk 1. The result of calling chunk2 from chunk1 And here’s the result. Escape the Land of LaTeX/Word for Statistical Reporting: The Ecosystem of R Markdown Webinar. Embedding RData files in Rmarkdown files for more reproducible analyses. For those of us interested in reproducible analysis, Rmarkdown is a great way of communicating our code to other researchers.
Rstudio, in particular, makes it very easy to create attractive HTML document containing text, code, and figures, which can then be sent to colleagues or put on the internet for anyone to see. “Mail merge” with RMarkdown. The term “mail merge” might not be familiar to those who have not worked in an office setting, but here is the Wikipedia definition: Mail merge is a software operation describing the production of multiple (and potentially large numbers of) documents from a single template form and a structured data source. The letter may be sent out to many “recipients” with small changes, such as a change of address or a change in the greeting line.Source: The other day I was working on creating personalized handouts for a workshop. Interactive documents: An incredibly easy way to use Shiny. How to set up your own R blog with Github pages and Jekyll Bootstrap.
This post is in reply to a request: How did I set up this R blog? Jupyter And R Markdown: Notebooks With R. When working on data science problems, you might want to set up an interactive environment to work and share your code for a project with others. You can easily set this up with a notebook. In other cases, you’ll just want to communicate about the workflow and the results that you have gathered for the analysis of your data science problem. R Markdown: How to number and reference tables. R Markdown is a great tool to make research results reproducible. However, in scientific research papers or reports, tables and figures usually need to be numbered and referenced. Unfortunately, R Markdown has no “native” method to number and reference table and figure captions.
The recently published bookdown package makes it very easy to number and reference tables and figures (Link). However, since bookdown uses LaTex functionality, R Markdown files created with bookdown cannot be converted into MS Word (.docx) files. Dynamic Documents with RMarkdown and Knitr. Getting started with R Markdown. Rmarkdown/pdf_document.R at master · rstudio/rmarkdown · GitHub. Fast-track publishing using the new R markdown – a tutorial and a quick look behind the scenes. The new R Markdown (rmarkdown-package) introduced in Rstudio 0.98.978 provides some neat features by combining the awesome knitr-package and the pandoc-system.
The system allows for some neat simplifications of the fast-track-publishing (ftp) idea using so called formats. I’ve created a new package, the Grmd-package, with an extension to the html_document format, called the docx_document. The formatter allows an almost pain-free preparing of MS Word compatible web-pages.
In this post I’ll (1) give a tutorial on how to use the docx_document, (2) go behind the scenes of the new rmarkdown-package and RStudio ≥ 0.98.978, (3) show what problems currently exists when skipping some of the steps outlined in the tutorial. Floating table of contents for your html reports using knitr. If you love knitr and rstudio and use them to produce long reports, you probably know that you can produce a table of contents in your html (and pdf) documents. In the newer rstudio (Version 0.98.801 or later) you do it by requesting a toc in the doc header, something like this.
R Markdown v2. Ly — Plot with ggplot2 and plotly within knitr reports. Plotly is a platform for making, editing, and sharing graphs. If you are used to making plots with ggplot2, you can call ggplotly() to make your plots interactive, web-based, and collaborative. For example, see plot.ly/~marianne2/166, shown below. Interactive Documents. Shiny - Gallery. Announcing RPubs: A New Web Publishing Service for R.