

Bioconductor - 3.3 Software Packages. Visualizing Concepts and Mappings - NCBO Wiki. With BioPortal, you can visualize ontology concepts and their mappings with other ontologies.
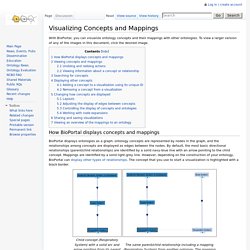
To view a larger version of any of the images in this document, click the desired image. How BioPortal displays concepts and mappings BioPortal displays ontologies as a graph: ontology concepts are represented by nodes in the graph, and the relationships among concepts are displayed as edges between the nodes. By default, the most basic directional relationships (parent/child relationships) are identified by a solid navy-blue line with an arrow pointing to the child concept. NCBO Widgets - NCBO Wiki. NCBO widgets are the HTML or Javascript code that you can put on your Web site or Web form to use BioPortal functionality there.
Using NCBO Widgets is just one of the ways in which you can use the NCBO technology directly on your Web site or in your application. Term-selection field on a form: You can add a text field to your Web form that will let users enter a term from a controlled vocabulary (e.g., terms from a single ontology) Example use case: Suppose you are running a tissue microarray database and users upload sample descriptions to your database using a web form. There is usually a field for the user to enter the diagnosis for the tissue sample that she is entering.
Usually, this field is a text-box or a drop down menu populated with a list of controlled terms. The free text-box is prone to errors, the drop-down gets too unwieldy with large terminologies. Silk - The Linked Data Integration Framework. Java and SPARQL. Jena Programming. BioMixer. Github Pages Links Latest Version Sample SNOMEDCT Concept Visualization Sample UBERON Concept Visualization Sample Ontology Overview for RDC Older Versions Sample SNOMEDCT Concept Visualization Sample UBERON Concept Visualization Sample Ontology Overview for RDC Node Collision Added, Property Edge Non-Overlap, Sample UBERON Arteries Concept Visualization Node Collision Removed, Node Charge Repulsion Increased, Property Edge Non-Overlap, Sample UBERON Arteries Concept Visualization.

National Center for Biomedical Ontology.
Vlab-cs-ucsb/SemRep: Semantic Differential Repair. Pillfill/rxnorm-client: An android/java library to use the NLM's RxNav/RxNorm Services. LexAccess. LexAccess Java 7.0, UTF-8 , 2013+ Release: A lexical access tool in the SPECIALIST Lexicon family is available.
This tool is called LexAccess. LexAccess provides access to information from the SPECIALIST lexicon. This tool is developed and released in Java since 2003 to integrate with the database in the new LexBuild system to provide the lexical-lookup capabilities not currently included within the lexical tools. Both plain text and XML formats of lexical records are available in LexAccess. A centralized web-based tool, LexBuild, was developed to build the SPECIALIST Lexicon in 2004. LexAccess takes various query options and returns lexical records. Lexical Tools. Lexical Tools Java 7.0, UTF-8 , 2016 Release: Lexical Tools is a set of fundamental core NLP tools for retrieving lexical variants.
Functions (flow components) include retrieving inflectional variants, uninflectional forms, spelling variants, derivational variants, synonyms, fruitful variants, normalization, UTF-8 to ASCII conversion, lowercase, etc.. There are over 62 flow components and 33 options offered in this tools set. This release includes the latest data integration from The SPECIALIST LEXICON along with completion of software change requests (SCRs). They are briefly described as follows. OntoFox. Indexing Initiative: Web API. This Java-based API to the Indexing Initiative Scheduler facility was created to provide users with the ability to programmatically submit jobs to the Scheduler Batch and Interactive facilities instead of using the web-based interface.

Three programs are accessible via the Web API: MetaMap, the NLM Medical Text Indexer (MTI), and SemRep. The functionality in these programs includes: automatic indexing of MEDLINE citations, concept-based query expansion, analysis of complex Metathesaurus strings, accurate identification of the terminology and relationships in anatomical documents, and the extraction of chemical binding relations from biomedical text. Whenever you see in the text below, it means that additional information is available by selecting the symbol. Semantic Knowledge Representation. Semantic Knowledge Representation. OntoFox. Swirrl. MeSH Browser - 2016. MeSH on Demand. MeSH on Demand identifies MeSH Terms in your text using the NLM Medical Text Indexer (MTI) program.
After processing, MeSH on Demand returns a list of MeSH Terms relevant to your text. For more information about MeSH on Demand, please see our NLM Technical Bulletin article. For electronic copies of recently presented posters on MeSH on Demand applications please send your requests to: NLMMESH-MOD@mail.nih.gov. TaskForces/CommunityProjects/LinkingOpenData/SemWebClients. TaskForces/CommunityProjects/LinkingOpenData/SemanticWebSearchEngines. This page collects links to Semantic Web Search Engines.
Semantic Web Search Engines use robots to crawl RDF data from the Web and provide search and navigation facilities over crawled data. The page is part of the community project SweoIG/TaskForces/CommunityProjects/LinkingOpenData Semantic Web Search Engines <sameAs.org> A search service that sits firmly in the Linked Data world - put in a Linked Data URI, and get back equivalent URIs. Differential Diagnosis. Ontobee SPARQL. RxMix. FilterByProperty Check the concept for a given property.
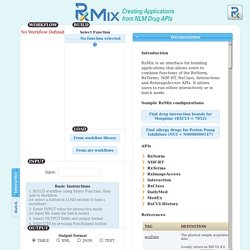
Returns the RXCUI if property was foundand it matches the value (if specified).INPUT: RXCUI; property_name; property_valueOUTPUT: RXCUI Check concept 892297 for SCHEDULE property value of 1 or 2 For more information click here. findRxcuiById Search by identifier to find RxNorm conceptsINPUT: identifier value; id_type (AMPID, ANDA, GCN_SEQNO, GFC, GPPC, HIC_SEQN, LISTING_SEQ_NO, MESH, MMSL_CODE, NDA, NDC, SNOMEDCT, SPL_SET_ID, UMLSCUI, UNII_CODE or VUID); allSourcesFlagOUTPUT: RXCUI Get the RXCUI from the UMLSCUI C0162723 For more information click here. findRxcuiByString. RxClass. BioMixer. Query repository · bio2rdf/bio2rdf-scripts Wiki. Ontorat. Ontorat is a web server that automatically generates and annotates ontology terms and axioms based on user-provided templates and specific design patterns.
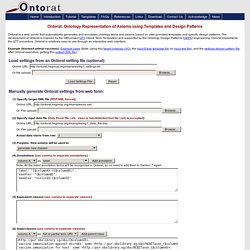
The development of Ontorat is inspired by the OBI project QTT (Quick Term Templates) and supported by the Ontology Design Patterns (ODPs) engineering. Ontorat implements the QTT procedure. Listing of 185 Ontology Building Tools. At the beginning of this year Structured Dynamics assembled a listing of ontology building tools at the request of a client. That listing was presented as The Sweet Compendium of Ontology Building Tools. Now, again because of some client and internal work, we have researched the space again and updated the listing [1]. All new tools are marked with <New> (new only means newly discovered; some had yet to be discovered in the prior listing). There are now a total of 185 tools in the listing, 31 of which are recently new, and 45 added at various times since the first release.
<Newest> reflects updates — most from the developers themselves — since the original publication of this post. Q&D SPARQL Browser. Marbles Linked Data Engine. News 2009/06/05: Added ANTLR properties in marbles.policy that prevented abstract view generation (thanks to Simon Reinhardt). 2009/05/28: Initial release of Marbles 1.0 on SourceForge. Contents 1. Concepts Views Apart from a full details view that lists all known properties for resource, Marbles allows for the generation of Fresnel-based views that are complemented by corresponding CSS stylesheets. There are currently two views in use, which were designed for DBpedia Mobile: The summary view features a short text describing the resource and optionally an image, a link to the resource's foaf:homepage and reviews from the Revyu rating site if existent. Q&D SPARQL Browser.