

RNA-Seq De novo Assembly Using Trinity. SSAKE — Canada's Michael Smith Genome Sciences Centre. SSAKE is a genomics application for assembling millions of very short DNA sequences.

Project Description The Short Sequence Assembly by K-mer search and 3' read Extension (SSAKE) is a genomics application for aggressively assembling millions of short nucleotide sequences by progressively searching for perfect 3'-most k-mers using a DNA prefix tree. SSAKE is designed to help leverage the information from short sequences reads by stringently clustering them into contigs that can be used to characterize novel sequencing targets.
*Best performance is achieved by quality-trimming your reads before assembly Enjoy SSAKE responsibly! Summary SSAKE is written in PERL and runs on Linux. Documentation René L Warren, Granger G Sutton, Steven JM Jones, Robert A Holt. 2007 (epub 2006 Dec 8). License Copyright (c) 2006-2011 Canada's Michael Smith Genome Science Centre. Credits René Warren, Granger Sutton, Steven Jones and Robert Holt All Releases. Applications - Whole Genome Sequencing : 454 Life Sciences, a Roche Company. Near-finished de novo assemblies of microbial genomes in days — Use the GS De Novo Assembler software to obtain a high-quality draft assembly in about 15 minutes, with minimal prior experience in bioinformatics.
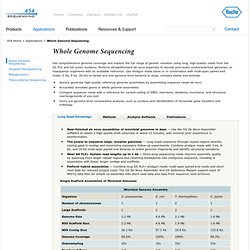
The power to sequence large, complex genomes — Long reads sequence through repeat regions directly, closing gaps in contigs and minimizing expensive follow-up experiments. Combine shotgun reads with 3 kb, 8 kb, and 20 kb multi-span paired end libraries to orient genomic fragments and identify structural variations. New! GS FLX+ System read lengths up to 1 kb — Extra-long sequencing reads improve assembly quality by spanning even larger repeat regions and resolving breakpoints into contiguous sequence, resulting in assemblies with fewer, longer contigs and scaffolds. Perform hybrid assemblies — Combine long GS FLX+ shotgun reads, multi-span paired end reads and short read data for reduced project costs.
Single Scaffold Assemblies of Microbial Genomes Shotgun Sequencing. SEQwiki. A Practical Comparison of De Novo Genome Assembly Software Tools for Next-Generation Sequencing Technologies. The advent of next-generation sequencing technologies is accompanied with the development of many whole-genome sequence assembly methods and software, especially for de novo fragment assembly.
Due to the poor knowledge about the applicability and performance of these software tools, choosing a befitting assembler becomes a tough task. Here, we provide the information of adaptivity for each program, then above all, compare the performance of eight distinct tools against eight groups of simulated datasets from Solexa sequencing platform. Considering the computational time, maximum random access memory (RAM) occupancy, assembly accuracy and integrity, our study indicate that string-based assemblers, overlap-layout-consensus (OLC) assemblers are well-suited for very short reads and longer reads of small genomes respectively. Short Oligonucleotide Analysis Package. The executable program and source code are now available on SourceForge: Other related tools(Know more): SOAPec_v2.01.tar.gz, a correction tool for SOAPdenovo: ErrorCorrection.tgz, another correction tool for SOAPdenovo: GapCloser-bin-v1.12-r6.tgz, a tool named GapCloser for SOAPdenovo: Data prepare module using assembled contig to do scaffold assembly: Please note that the old version of SOAPdenovo is out of maintainance now, if you want to achieve the old version of SOAPdenovo, please contact us (xieyl@genomics.cn or bgi-soap@googlegroups.com).
Publication Update Log 1) Source code of 63mer and 127mer versions were merged. Installation. Research Laboratory Geneva - Edena: very short reads assembler. Edena v3: de novo short reads assembler References De novo bacterial genome sequencing: millions of very short reads assembled on a desktop computer.

Hernandez D, François P, Farinelli L, Osterås M, Schrenzel J. Genome Research. 18:802-809, 2008. De novo finished 2.8 Mbp Staphylococcus aureus genome assembly from 100 bp short and long range paired-end reads. Announcement list To get informed about new releases, you are welcome to subscribe to the Edena announce list by entering your email address below. Contact Dr David HERNANDEZ Download Edena V3.131028 (October 28, 2013) Minor bugs fix Some speed optimizations Better errors checking Edena V3.130110 (January 11, 2013) Updated the interactive shell. EdenaV3.121122 (November 27, 2012) Velvet: a sequence assembler for very short reads. 29/03/2011: Velvet 1.1 Velvet is now multithreaded, thanks to the use of the OMP library. 15/06/2010: Velvet 1.0 The Columbus module within Velvet now allows the assembly process to be assisted by alignment information onto a set of reference sequences, for example for assisted transcriptome assembly, or local re-sequencing of structural variants. 22/12/2009: Pebble and Rock Band paper in PLoS ONE The description of the inner workings of Pebble and RockBand (i.e. the paired-end and mixed-length modules) is now available in PLoS ONE 09/09/2008: Velvet 0.7 This release contains the new Pebble algorithm which now replaces the BreadCrumb paired-end module.
