

Bioinformatics, Comparative Genomics and Molecular Evolution. Pandoravirus: missing link discovered between viruses and cells. Researchers at IGS, the genomic and structural information laboratory (CNRS/Aix-Marseille University), working in association with the large-scale biology laboratory (CEA/Inserm/Grenoble Alpes University) have just discovered two giant viruses which, in terms of number of genes, are comparable to certain eukaryotes, microorganisms with nucleated cells.
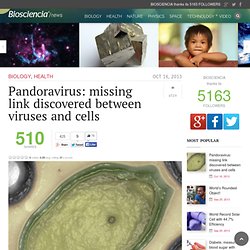
The two viruses – called “Pandoravirus” to reflect their amphora shape and mysterious genetic content – are unlike any virus discovered before. This research appeared on the front page of Science on July 19, 2013. With the discovery of Mimivirus ten years ago and, more recently, Megavirus chilensis, researchers thought they had reached the farthest corners of the viral world in terms of size and genetic complexity. Norine. NORINE is a platform that includes a database of nonribosomal peptides together with tools for their analysis.
Norine currently contains more than 1000 peptides. The name Norine stands for Nonribosomal peptides, with ine as a typical ending of peptide names. For each peptide, the database stores its structure as well as various annotations such as the biological activity, producing organisms, bibliographical references among others. In the Pipeline: Perspectives on the Development and Commercialization of Molecular Biomarkers.
Broad Institute of MIT and Harvard. Norine. Publications - Technology - Oxford Nanopore Technologies. Search. GMOD. Overview. ... formerly titled "GMOD for the Biologist".
This page provides an overview of the GMOD project. It does not assume any particular background in computing. Introduction. Biopackages HOWTO. This page or section needs to be edited.
Please help by editing this page to add your revisions or additions. RPM-based Linux distributions, including Fedora Core and CentOS, can install GMOD software (chado databases, GBrowse, GMODWeb, etc) using the RPMs located at These can be installed using the yum installer tool. This process is an easy and quick way to install GMOD tools. The only requirement is that you use one of the supported distributions (Centos4 is the supported & tested platform for GMOD) and you install biopackages on a clean system (e.g. no source installed software). We recommend you do not mix source and RPM software installs unless you really know what you are doing since they could (silently) conflict with each other. At the time of this writing (April 2007) complete RPMs for Fedora Core 2, 5, and CentOS 4 are available, and supported architectures included 32- and 64-bit Intel platforms.
Biopackages.net hosts a yum repository for distribution of Linux packages. Phylogenetics. Evolutionary Relationships Between Maize & Other Grass Species Maize is a member of the grass family Poaceae (Gramineae), a classification it shares with many other important agricultural crops, including wheat (Triticum aevestium), rice (Orzya sativa), oats (Avena), sorghum (Sorghum), barley (Hordeum) and sugarcane (Saccharum).

Based on fossil evidence, it is estimated that these major grass lineages arose from a common ancestor within the last 55-70 million years, near the end of the reign of dinosaurs. Maize is further organized in genus Zea, a group of annual and perennial grasses native to Mexico and Central America. Genus Zea includes the wild taxa, known collectively as “teosinte” (Zea ssp.), as well as domesticated corn, or “maize” (Zea mays ssp. mays). Grass Relationships: Tracing the Phylogeny of the Hexaploid OatAvena sativawith Satellite DNAs. Recombinant Protein Expression, Virus Like Particles, baculovirus expression system. Phylogenetics. Using Bioconductor to Analyze your 23andme Data. Bioconductor is one of the open source projects of which I am most fond.

The documentation is excellent, the community wonderful, the development fast-paced, and the software very well written. There’s a new package in the development branch (due to be released as 2.10 very soon) called gwascat. gwascat is a package that serves as an interface to the NHGRI’s database of genome-wide association studies. Loading the package with library(gwascat) creates a GRanges instance of SNPs and their diseases. GRanges is a fundamental data structure in Bioconductor (specifically the GenomicRanges package) that is designed to hold ranges on genomes efficiently, as well as metadata about the ranges. In this case, the object gwrngs holds SNP ranges (well, locations) and metadata provided by the GWA studies in NHGRI’s database. While I really do like 23andme’s interface to one’s genotype information and research, the gwascat package offers some nice data mining power. Dear Oxford Nanopore: How About Some Data? On the Preservation of Published Bioinformatics Code on Github « I wish you'd made me angry earlier.
A few months back I posted a quick analysis of trends in where bioinformaticians choose to host their source code.
A clear trend emerging in the bioinformatics community is to use github as the primary repository of bioinformatics code in published papers. While I am a big fan of github and I support its widespread adoption, in that post I noted my concerns about the ease with which an individual can delete a published repository. In contrast to SourceForge, where it is extremely difficult to delete a repository once files have been released and this can only be done by SourceForge itself, deleting a repository on github takes only a few seconds and can be done (accidentally or intentionally) by the user who created the repository. Just to see how easy this is, I’ve copied the process for deleting a repository on github here: Go to the repo’s admin page Click “Delete this repository”