


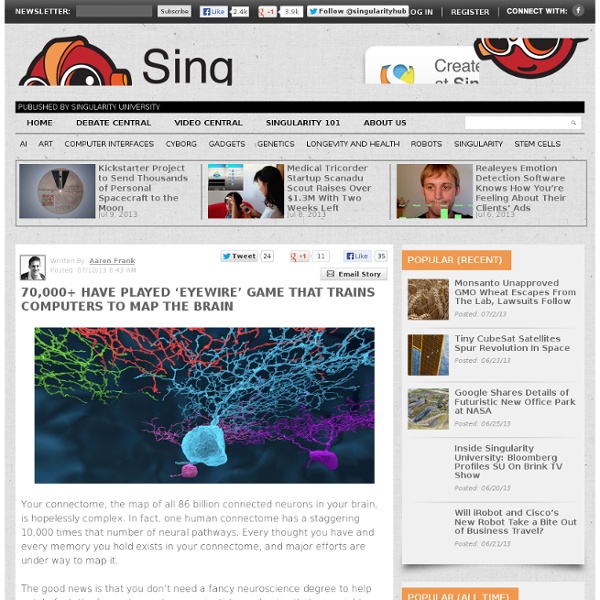
Sequencing the Connectome Converting connectivity into a sequencing problem can be broken down conceptually into three components. (A) Label each neuron with a unique sequence of nucleotides — a DNA “barcode.” (B) Associate barcodes from synaptically connected neurons with one another, so that each neuron can be thought of as a “bag of barcodes” — copies of its own “host” barcode and copies of “invader” barcodes from synaptic partners. (C) Join host and invader barcodes into barcode pairs. These pairs can be subjected to high-throughput sequencing. (Credit: Anthony M. A team of neuroscientists led by Professor Anthony Zador, Ph.D., of Cold Spring Harbor Laboratory have proposed a revolutionary new way to create a connectivity map (“connectome”) of the whole brain of the mouse at the resolution of single neurons: high-throughput DNA sequencing. This reconstruction of serial electron micrographs has yielded what to date is the only complete connectome, that of C. elegans (a nemotode or roundworm).
Cosmos: A Space-Time Odyssey: Carl Sagan’s show updated with Neil Tyson. Photo by Fox, form the video In 1980, Carl Sagan changed the face of science forever. In that year, PBS broadcast the TV series Cosmos: A Personal Voyage. Sagan’s words and voice drove the show, taking the 500 million people who watched with him as he showed us the Universe, from the distant reaches of its redshifted expansion to the chemical processes as our brains create our minds. As Sagan himself said, “We are a way for the cosmos to know itself.” That show transformed how people saw science and also how scientists informed the public. My old friend and brilliant science communicator Neil Tyson is doing just that. I prefer not to prejudge a show too much on the trailer, but it’s hard to resist a few comments. I’m very much looking forward to this. Photo by Phil Plait (well, Neil actualy took it but it was my phone). I’ll note that some people have their doubts about this because Seth MacFarlane is involved. Photo by Phil Plait
The Brain CONNECT Project lifehacker Brain Research through Advancing Innovative Neurotechnologies (BRAIN) NIH Home > Research & Training What is the BRAIN Initiative? The Brain Research through Advancing Innovative Neurotechnologies (BRAIN) Initiative is part of a new Presidential focus aimed at revolutionizing our understanding of the human brain. By accelerating the development and application of innovative technologies, researchers will be able to produce a revolutionary new dynamic picture of the brain that, for the first time, shows how individual cells and complex neural circuits interact in both time and space. Long desired by researchers seeking new ways to treat, cure, and even prevent brain disorders, this picture will fill major gaps in our current knowledge and provide unprecedented opportunities for exploring exactly how the brain enables the human body to record, process, utilize, store, and retrieve vast quantities of information, all at the speed of thought. A map of overall task-fMRI brain coverage from the seven tasks used in the Human Connectome Project. Meeting Information
Navy Researchers Put Dark Lightning to the SWORD Want to stay on top of all the space news? Follow @universetoday on Twitter Dark lightning occurs within thunderstorms and flings gamma rays and antimatter into space. (Science@NASA video) Discovered “by accident” by NASA’s Fermi Gamma-ray Space Telescope in 2010, dark lightning is a surprisingly powerful — yet invisible — by-product of thunderstorms in Earth’s atmosphere. What’s more, these gamma-ray outbursts originate at relatively low altitudes well within the storm clouds themselves. Terrestrial Gamma-ray Flashes (TGFs) are extremely intense, sub-millisecond bursts of gamma rays and particle beams of matter and anti-matter. These models also suggest that the particle beams are intense enough to distort and collapse the electric field within thunderstorms and may, therefore, play an important role in regulating the production of visible lightning. A team of NRL Space Science Division researchers, led by Dr. As a next step, Dr. Simulation of a Boeing 737 struck by dark lightning.
List of animals by number of neurons This is a list of representative animals by the number of neurons in their whole nervous system and the number of neurons in their brain (for those with a brain). These numbers are estimates derived by multiplying the density of neurons in a particular animal by the average volume of the animal's brain. Overview[edit] Neurons may be packed to form structures such as the brain of vertebrates or the neural ganglions of insects. The number of neurons and their relative abundance in different parts of the brain is a determinant of neural function and, consequently, of behavior. Whole nervous system[edit] Cerebral cortex[edit] See also[edit] References[edit]
Creatures Lived On Land 2.2 Billion Years Ago, New Evidence Suggests South African fossils push the rise of oxygen and life on land to hundreds of millions of years earlier than previously thought. Researchers at the University of Oregon have recently unearthed fossils in South Africa that present evidence of life on land 2.2 billion years ago—four times older than traditionally thought. The fossilized organisms, dubbed Diskagma buttonii, are no bigger than the size of a standard match head and were found threaded together in bunches. Though researchers are still unsure as to their biological function, the organisms most closely resemble a modern fungus called Geosiphon . The results of the study, led by geologist Gregory J. "There is independent evidence for cyanobacteria, but not fungi, of the same geological age, and these new fossils set a new and earlier benchmark for the greening of the land," Retallack says in a statement . “At last we have an idea of what life on land looked like in the Precambrian," Retallack says.
List of topics related to brain mapping The following is a list of topics related to brain mapping, and major brain mapping research projects (listed below). Coverage is intended to be broad and comprehensive, and adequately cover the entire brain mapping field. Topics included are in rough proportion to their generally accepted overall importance to the human brain structure and function. It is not intended to be recursively exhaustive in every possible direction but to give an overview of what areas of knowledge may be impacted by the large new brain mapping research initiatives. While the emphasis here is on physical brain structure, functional aspects are also included. Mind concepts (as in mind vs. body), and cognitive and behavioral aspects, are introduced where they have at least a fairly direct connection to physical aspects of the brain, neurons, spinal cord, nerve networks, neurotransmitters, etc. Topics are roughly clustered as shown in the table of contents. Broad Scope[edit] The Neuron doctrine[edit] General[edit]
Could Life Be Older Than Earth Itself? Applying a maxim from computer science to biology raises the intriguing possibility that life existed before Earth did and may have originated outside our solar system, scientists say. Moore’s Law is the observation that computers increase exponentially in complexity, at a rate of about double the transistors per integrated circuit every very two years. If you apply Moore’s Law to just the last few years’ rate of computational complexity and work backward, you’ll get back to the 1960s, when the first microchip was, indeed, invented. Now, two geneticists have applied Moore’s Law to the rate at which life on Earth grows in complexity — and the results suggest organic life first came into existence long before Earth itself. ANALYSIS: Clues of Life's Origins Found in Galactic Cloud The results suggest life first appeared about 10 billion years ago, far older than the Earth’s projected age of 4.5 billion years. Sharov and Gordon’s idea raises other intriguing possibilities.
BRAIN Initiative Understanding how the brain works is arguably one of the greatest scientific challenges of our time. The BRAIN Initiative (Brain Research through Advancing Innovative Neurotechnologies, also referred to as the Brain Activity Map Project) is a proposed collaborative research initiative announced by the Obama administration on April 2, 2013, with the goal of mapping the activity of every neuron in the human brain.[2][3][4][5][6] Based upon the Human Genome Project, the initiative has been projected to cost more than $300 million per year for ten years.[2] Announcement[edit] Experimental approaches[edit] News reports said the research would map the dynamics of neuron activity in mice and other animals[3] and eventually the tens of billions of neurons in the human brain.[8] Working group[edit] The advisory committee is:[12] [edit] Scientists offered differing views of the plan. The projects face great logistical challenges. See also[edit] References[edit] External links[edit] BRAIN Initiative website
Quantum Computer to Log Onto Quantum Internet When it comes to data crunching, quantum computers will leave today's fastest processors in the dust. For starters, a quantum computer would be able to store more bits of information in its memory than there are particles in the universe. And where a conventional silicon-based computer handles one computation at a time in sequence, a quantum computer would work on millions at once. That kind of staggering power would give a single quantum computer the ability to simulate a whole world in a holographic environment, replicate biological systems to understand diseases and find cures, solve the loads of equations necessary to create extremely accurate weather forecasting and simulate how subatomic particles interact, showing fundamentally how everything in the universe works. NEWS: Quantum of Solar: Cells Get Mystery Power Boost In recent months, different groups of scientists and engineers have made important strides toward this amazing new world.
Human Connectome Project The Human Connectome Project (HCP) is a five-year project sponsored by sixteen components of the National Institutes of Health, split between two consortia of research institutions. The project was launched in July 2009[1] as the first of three Grand Challenges of the NIH's Blueprint for Neuroscience Research.[2] On September 15, 2010, the NIH announced that it would award two grants: $30 million over five years to a consortium led by Washington University in Saint Louis and the University of Minnesota, and $8.5 million over three years to a consortium led by Harvard University, Massachusetts General Hospital and the University of California Los Angeles.[3] The goal of the Human Connectome Project is to build a "network map" that will shed light on the anatomical and functional connectivity within the healthy human brain, as well as to produce a body of data that will facilitate research into brain disorders such as dyslexia, autism, Alzheimer's disease, and schizophrenia.[4]
Insecticides Spreading to Wildflowers Poisons Bees In a recent study, pollen contaminated with insecticides and fungicides poisoned honey bees and weakened the bees’ resistance to a deadly parasite. What’s more, the poisoned pollen didn’t just come from agricultural crops. In fact, many bees collected most of their pollen from wildflowers, as opposed to the fields that farmers pay beekeepers to pollinate. Previous studies have mostly focused on insecticides, particularly neonicotinoids, as a cause of the widespread death of honey bees (Apis mellifera) due to a mysterious syndrome, known as colony collapse disorder. In December, the European Union will implement a ban on three types of neonicotinoid insecticide, known as clothianidin, imidacloprid and thiametoxam, reported the BBC. NEWS: Bee Decline Threatens Entire Ecosystems However, the new study, published in PLOS ONE, suggests that banning neonicotinoids might not be enough to halt the disappearance of the bees. ANALYSIS: 25,000 Dead Bees in Target Store Parking Lot