


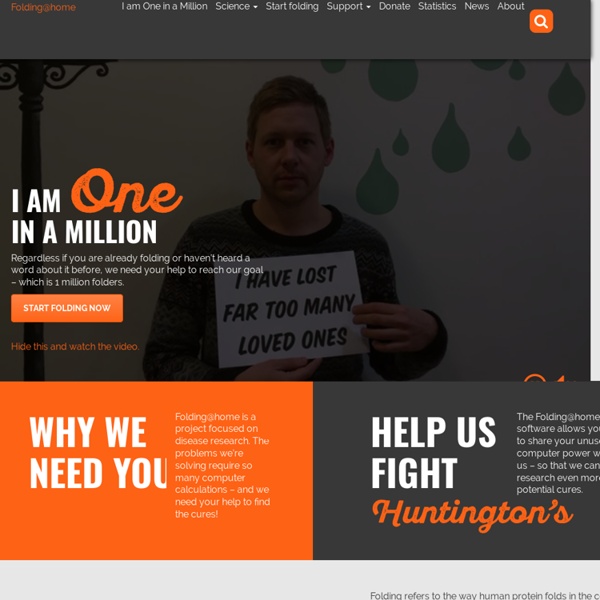
Folding@home Distributed computing project simulating protein folding Folding@home (FAH or F@h) is a distributed computing project aimed to help scientists develop new therapeutics for a variety of diseases by the means of simulating protein dynamics. This includes the process of protein folding and the movements of proteins, and is reliant on simulations run on volunteers' personal computers.[5] Folding@home is currently based at the University of Pennsylvania and led by Greg Bowman, a former student of Vijay Pande.[6] The project utilizes graphics processing units (GPUs), central processing units (CPUs), and ARM processors like those on the Raspberry Pi for distributed computing and scientific research. Folding@home is one of the world's fastest computing systems. Background[edit] Examples of application in biomedical research[edit] Alzheimer's disease[edit] Alzheimer's disease is linked to the aggregation of amyloid beta protein fragments in the brain (right). Huntington's disease[edit] Cancer[edit]
Education World ® : Curriculum: Journal Writing Every Day: Teachers Say It Really Works! One of the best things about daily journal writing is that it can take so many forms. Teachers can use journal writing to meet specific goals, or the purpose can be wide open. Some teachers check journal writing and work on polishing skills; others use journals as the one "uncorrected" form of writing that students produce. Some teachers provide prompts to help students begin their writing. Others leave decisions about the direction and flow of student journals up to the students. This week, Education World talked with teachers who use daily journal writing in their classrooms. "They have come such a long way in their writing," said teacher Laura Black. Daily journal writing has helped Black's students at St. "They are not afraid to take on any writing that may come their way," added Black, "because they have built up extreme confidence." That's progress any teacher would be proud of -- and Laura Black teaches first grade! At St. Kathy Thomson teaches at S.
2011 About.com Readers' Choice Awards Winners - About Web Browsers - Best Privacy/Security Add-On Privacy and security while browsing the Web is important to all of us, as evidenced by the fervent voting in this category. The five finalists featured an impressive selection of tools intended to make everday life on the Web safer. After more than three weeks of non-stop action, the readers have made their decision. The reigning champion in the 2011 Best Privacy/Security Add-On category, for the second year in a row, is NoScript!Final Voting Results*NoScript - 56%WOT (Web of Trust) - 33%BetterPrivacy - 4%LastPass - 3%FlashBlock - 2%*denotes winner 2010 Winner: NoScript
Ideas to build community - Activism Why is community building an important part of activism? Without community we are alone. We build communities of friends, neighbours, relatives and co-workers. By strenghtening these communities we make stronger bonds with those around us. smile at the people you pass on the street, sit next to on public transit etc. have a potluck with your neighbours, friends, co-workers or relatives turn off your TV (you will have more time to be with your community) have a neighbour clean-up of trash start a item exchange (clothes, furniture, electronics etc) start a child-minding co-op start a community kitchen volunteer your time at a retirement/rest home start a community garden walk your dog with your neighbour meet your neighbours (invite them for tea, bring them some baked goods, say *hi* to them on the street) look up when you walk, and greet people support your local economy by buying from local merchants, producers and artisans share your skills with others
GPUGRID.net GPUGRID is a distributed computing project hosted by Pompeu Fabra University and running on the Berkeley Open Infrastructure for Network Computing (BOINC) software platform. It performs full-atom molecular biology simulations that are designed to run on Nvidia's CUDA-compatible graphics processing units. Former support for PS3s[edit] See also[edit] List of distributed computing projects References[edit] Further reading[edit] External links[edit]
Campbell Center for Training in Preventive Collections Care, Museum Studies, Historic Preservation, and Conservation The Doomsday Clock from the Bulletin of Atomic Scientists tackles biotechnology SCOTT OLSON/AFP/Getty Images Shortly after the end of World War II, Albert Einstein, referring to the new global danger of nuclear weapons, uttered his now famous warning: “Everything has changed, save the way we think.” Accordingly, he and Robert Oppenheimer established the Bulletin of the Atomic Scientists to help warn the public about the dangers of nuclear war. Perhaps the most visible face of the bulletin—for which I am currently co-chair of the board of sponsors—is the “Doomsday Clock.” In total, the clock has been adjusted 20 times, moving as close to two minutes to midnight in 1953, after the United States and Soviet Union each first tested thermonuclear devices, and as far as 17 minutes to midnight in 1991, after the United States and Soviet Union signed the Strategic Arms Reduction Treaty. These developments are thrilling for scientists and technologists who love to take things apart and put them back together.
Dreamer - Search the Web, Change the World