

Synthetic Biology:BioBricks/3A assembly. From OpenWetWare Introduction 3A assembly (which stands for three antibiotic assembly) is a method for assembling two BioBrick parts together.
It differs from the more common assembly approach and the Silver lab approach in that it relies on three way ligation (between the two parts and the backbone vector) for assembly rather than two way ligation between a part and a part + vector molecule. It uses both positive and negative selection to reduce/eliminate the number of incorrect assemblies that give rise to colonies after transformation.
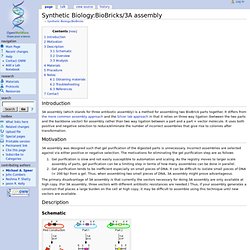
Motivation 3A assembly was designed such that gel purification of the digested parts is unnecessary. Gel purification is slow and not easily susceptible to automation and scaling. The primary disadvantage of 3A assembly is that currently the vectors necessary for doing 3A assembly are only available at high copy. Description Schematic Schematic of 3A assembly The red plasmid is the construction vector.
Overview Analysis Materials Procedure Notes Troubleshooting. Synthetic Biology:BioBricks/Standard FACS protocol. From OpenWetWare MIT Synthetic Biology Working GroupStandardized FACS ProtocolDraft 1 – 9/20/04 This protocol sets out to standardize the components of a FACS experiment necessary to allow repeatable and comparable results.
The output of these experiments is typically fluorescence from a genetic regulatory network. Media and Glassware Provisionally, supplemented M9 media will be used for all cultures. 1x M9 Minimal Salts (from Sigma): 12.8 g/L NaHPO4.7H20 3.0 g/L KH2PO4 0.5 g/L NaCl 1.0 g/L NH4Cl 0.1mM MgSO4 2mM CaCl2 0.4% Glucose/Glycerol 0.1% Casamino Acids Cultures should, where possible, be grown in flasks rather than tubes.
Strains and Plasmids It is unrealistic to specify one strain and plasmid for all characterization work but initial discussion suggests that, where possible, MC4100 and a 3-series plasmid should be used. Protocol. Synthetic Biology:BioBricks/Part fabrication. From OpenWetWare This page is intended as a how-to guide for constructing novel BioBrick parts for submission to the Registry of Standard Biological Parts.
For those already familiar with making BioBrick parts, see the quick reference guide. For help in assembling two preexisting BioBricks parts together, see one of the following pages. Introduction This page serves as documentation for how to construct a BioBrick part. The exact approach used when fabricating a BioBrick part depends on the fabrication method (PCR or direct synthesis) as well as the type of part being constructed (standard part or protein coding sequence). Constructing a BioBrick part via PCR A BioBrick can be constructed via PCR if there already exists template DNA from which the BioBricks can be amplified or if the part is short enough that it can be created by primer annealing and extension. Standard part fabrication Use this approach for promoters, ribosome binding sites, terminators and most other BioBricks parts. Prefix. BioBricks construction tutorial. From OpenWetWare Contacts Barry Canton, Jason Kelly, Will Bosworth.
Silver: BB Strategy. Silver: Protocols.