

Foundations for the design and implementation of synthetic genetic circuits : Abstract : Nature Reviews Genetics. Efficient and sequence-independent ... [Proc Natl Acad Sci U S A. 2012. Foundations for the design and implementation of synthetic genetic circuits : Abstract : Nature Reviews Genetics. Natural strategies for the spatial optimizatio... [Nat Chem Biol. 2012. Natural strategies for the spatial optimizatio... [Nat Chem Biol. 2012. Rapid and orthogonal logic gating with a gibbe... [Nat Chem Biol. 2012. Rapid and orthogonal logic gating with a gibbe... [Nat Chem Biol. 2012. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Reprogramming the Genetic Code. Synthetic Biology: Mapping the Scientific Landscape. Results. Synthetic Genetic Polymers Capable of Heredity and Evolution. Synthetic Genetic Polymers Capable of Heredity and Evolution.
Mapping the Environmental Fitness Landscape of a Synthetic Gene Circuit. Abstract Gene expression actualizes the organismal phenotypes encoded within the genome in an environment-dependent manner.
Among all encoded phenotypes, cell population growth rate (fitness) is perhaps the most important, since it determines how well-adapted a genotype is in various environments. Traditional biological measurement techniques have revealed the connection between the environment and fitness based on the gene expression mean. Yet, recently it became clear that cells with identical genomes exposed to the same environment can differ dramatically from the population average in their gene expression and division rate (individual fitness). For cell populations with bimodal gene expression, this difference is particularly pronounced, and may involve stochastic transitions between two cellular states that form distinct sub-populations.
Author Summary It is common belief that the properties of cells depend on their environment and on the genes they carry. Trends in Biotechnology - Distributed computation: the new wave of synthetic biology devices. Rationally designed families of orthogonal RNA... [Nat Chem Biol. 2012. A Biobrick Library for Cloning Custom Eukaryotic Plasmids. Researchers often require customised variations of plasmids that are not commercially available.

Here we demonstrate the applicability and versatility of standard synthetic biological parts (biobricks) to build custom plasmids. For this purpose we have built a collection of 52 parts that include multiple cloning sites (MCS) and common protein tags, protein reporters and selection markers, amongst others. Importantly, most of the parts are designed in a format to allow fusions that maintain the reading frame. We illustrate the collection by building several model contructs, including concatemers of protein binding-site motifs, and a variety of plasmids for eukaryotic stable cloning and chromosomal insertion. For example, in 3 biobrick iterations, we make a cerulean-reporter plasmid for cloning fluorescent protein fusions. Figures Citation: Constante M, Grünberg R, Isalan M (2011) A Biobrick Library for Cloning Custom Eukaryotic Plasmids.
MIT-CSAIL-TR-2012-008. Balanced Codon Usage Optimizes Eukaryotic Translational Efficiency. Abstract Cellular efficiency in protein translation is an important fitness determinant in rapidly growing organisms.
It is widely believed that synonymous codons are translated with unequal speeds and that translational efficiency is maximized by the exclusive use of rapidly translated codons. Here we estimate the in vivo translational speeds of all sense codons from the budding yeast Saccharomyces cerevisiae. Surprisingly, preferentially used codons are not translated faster than unpreferred ones.
We hypothesize that this phenomenon is a result of codon usage in proportion to cognate tRNA concentrations, the optimal strategy in enhancing translational efficiency under tRNA shortage. Author Summary Although an amino acid can be encoded by multiple synonymous codons, these codons are not used equally frequently in a genome. Figures Citation: Qian W, Yang J-R, Pearson NM, Maclean C, Zhang J (2012) Balanced Codon Usage Optimizes Eukaryotic Translational Efficiency.
Current Opinion in Biotechnology - A retrosynthetic biology approach to therapeutics: from conception to delivery. Engineering Molecular Circuits Usin... [Annu Rev Chem Biomol Eng. 2012. Integrated Electromicrobial Conversion of CO2 to Higher Alcohols. 1203808109.full. Rationally designed families of orthogonal RNA... [Nat Chem Biol. 2012. A Sensitive Switch for Visualizing Natural Gene Silencing in Single Cells - ACS Synthetic Biology. † School of Biological and Health Systems Engineering, Arizona State University, Tempe, Arizona 85287, United States ‡ Laboratory of Cellular and Molecular Engineering, University of Bologna, I-47521 Cesena, Italy § Department of Systems Biology, Harvard Medical School, Boston, Massachusetts 02115, United States The Wyss Institute for Biologically Inspired Engineering, Harvard Medical School, Boston, Massachusetts 02115, United States ACS Synth.
Biol., 2012, 1 (3), pp 99–106 DOI: 10.1021/sb3000035. Signaling-mediated bacterial persister formation. [Nat Chem Biol. 2012. Signaling-mediated bacterial persister formation. [Nat Chem Biol. 2012. Generation of a Synthetic Memory Trace. The Imaginary Mind of a Mouse. Rational Diversification of a Promoter Providing Fine-Tuned Expression and Orthogonal Regulation for Synthetic Biology. Yeast is an ideal organism for the development and application of synthetic biology, yet there remain relatively few well-characterised biological parts suitable for precise engineering of this chassis.
In order to address this current need, we present here a strategy that takes a single biological part, a promoter, and re-engineers it to produce a fine-graded output range promoter library and new regulated promoters desirable for orthogonal synthetic biology applications. A highly constitutive Saccharomyces cerevisiae promoter, PFY1p, was identified by bioinformatic approaches, characterised in vivo and diversified at its core sequence to create a 36-member promoter library. TetR regulation was introduced into PFY1p to create a synthetic inducible promoter (iPFY1p) that functions in an inverter device. Figures Editor: Mick F. Received: December 20, 2011; Accepted: February 13, 2012; Published: March 19, 2012 Copyright: © 2012 Blount et al. Introduction. 1108.3752. Phase transitions in the assembly of multivalent sign... [Nature. 2012. A Logic-Gated Nanorobot for Targeted Transport of Molecular Payloads.
An Engineered Microbial Platform for Direct Biofuel Production from Brown Macroalgae. Synthetic biology: Simplifying design. [Nat Rev Genet. 2012. A sensing array of radically coupled genetic 'biopixels' [Nature. 2011. Synthetic quorum-sensing circuit to control consortial biofilm formation and dispersal in a microfluidic device : Nature Communications. Microfluidic biofilm engineering circuit The microfluidic biofilm engineering (μBE) signalling circuit was constructed in E. coli using two engineered biofilm-dispersing proteins, Hha13D6 (ref. 15) and BdcAE50Q16, along with the P. aeruginosa LasI/LasR QS system (Fig. 1a) for use in the novel microfluidic device (Fig. 1b).
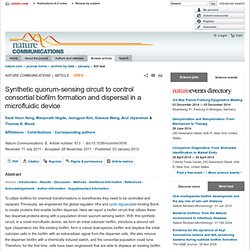
E. coli hha31 was used as the host as deletion of hha increases biofilm formation14 and provides a background in which there is no wild-type Hha. Lactococcal promoter CP2532 was used as the strong constitutive promoter for two of the three proteins on each plasmid. To obtain high concentrations of intercellular signal 3oC12HSL and regulator LasR, a synthetic ribosomal-binding site (RBS II)32 was first utilized. However, high expression of lasI or lasR was deleterious; thus, we used the native RBS of these genes. (a) The two E. coli cell types communicate by using the LasI/LasR QS module of P. aeruginosa.
Model-Driven Engineering of RNA Devices to Quantitatively Program Gene Expression. Model-Driven Engineering of RNA Devices to Quantitatively Program Gene Expression. Emerging biomedical applications of synthetic biology : Abstract : Nature Reviews Genetics. In silico feedback for in vivo regulation of a gene expression circuit : Nature Biotechnology. We show that difficulties in regulating cellular behavior with synthetic biological circuits may be circumvented using in silico feedback control.

By tracking a circuit's output in Saccharomyces cerevisiae in real time, we precisely control its behavior using an in silico feedback algorithm to compute regulatory inputs implemented through a genetically encoded light-responsive module. Moving control functions outside the cell should enable more sophisticated manipulation of cellular processes whenever real-time measurements of cellular variables are possible. View full text Figures. From DNA to Targeted Therapeutics: Bringing Synthetic Biology to the Clinic. Science Translational Medicinestm.sciencemag.org Sci Transl Med 26 October 2011: Vol. 3, Issue 106, p. 106ps42 Sci.
Transl. Med. DOI: 10.1126/scitranslmed.3002944 Perspective Bioengineering + Author Affiliations ↵*Correspondence. Synthetic biology aims to make biological engineering more scalable and predictable, lowering the cost and facilitating the translation of synthetic biological systems to practical applications. Copyright © 2011, American Association for the Advancement of Science. Bayesian design of synthetic biological systems. Engineering modular and orthogonal genetic logic gates for robust digital-like synthetic biology : Nature Communications. Circuit design and control components characterization The AND gate (Fig. 1; Supplementary Fig.

S1) comprises two co-activating genes hrpR and hrpS and one σ54-dependent hrpL promoter, and can integrate two interchangeable environmental signal inputs to generate one interchangeable output. Arming yeast for synthetic biology : Nature Biotechnology. Sequential Establishment of Stripe Patterns in an Expanding Cell Population. 110118_imaginenano. NanoBioMed2011_Douglas_Shawn_shawn.douglas. Yeast thrives with partially synthetic genome. Entrainment of a Population of Synthetic Genetic Oscillators.
Multi-Input RNAi-Based Logic Circuit for Identification of Specific Cancer Cells. Chemical and Genetic Engineering of Selective Ion Channel–Ligand Interactions. A Grand Challenge in Biology. Science Magazine: Sign In. How synthetic can “synthetic biology” be?

A literal interpretation of the name of this new life science discipline invokes expectations of the systematic construction of biological systems with cells being built module by module—from the bottom up. But can this possibly be achieved, taking into account the enormous complexity and More. Science Magazine: Sign In. Science Magazine: Sign In. Synthetic biology is an emerging field focused on engineering biomolecular systems and cellular capabilities for a variety of applications.
Substantial progress began a little over a decade ago with the creation of synthetic gene networks inspired by electrical engineering. Since then, the field has designed and built increasingly complex circuits More. Science Magazine: Sign In. A major goal of synthetic biology is to develop a deeper understanding of biological design principles from the bottom up, by building circuits and studying their behavior in cells.
Investigators initially sought to design circuits “from scratch” that functioned as independently as possible from the underlying cellular system. More recently, More A major goal of synthetic biology is to develop a deeper understanding of biological design principles from the bottom up, by building circuits and studying their behavior in cells. Investigators initially sought to design circuits “from scratch” that functioned as independently as possible from the underlying cellular system. More recently, researchers have begun to develop a new generation of synthetic circuits that integrate more closely with endogenous cellular processes.
Science Magazine: Sign In. Science Magazine: Sign In. Science Magazine: Sign In. Science Magazine: Sign In. The Allure of Synthetic Biology. Biologists have been manipulating genomes ever since Paul Berg first described a method to covalently join duplex DNA molecules in 1972. Despite key fundamental insights, a thriving biotechnology industry, and a growing number of medical applications, there have been limits to what has been possible.
Now, synthetic biology goes beyond engineering individual genes to the construction of DNA-encoded circuits that can be programmed to control cell behavior. This emerging field brings together biologists, physicists, chemists, and engineers who seek both to understand life and to build new biological functions. For example, Harvard's George Church wants to redesign the genetic code (p. 1236). The potential of synthetic biology has also attracted artists who want to critique it and make use of its techniques (p. 1242), as well as do-it-yourself biologists, some of whom have set up community labs (p. 1240). Expanding the Genetic Code of Escherichia coli with Phosphoserine.
Automatic Compilation from High-Level Biologically-Oriented Programming Language to Genetic Regulatory Networks. Citation: Beal J, Lu T, Weiss R (2011) Automatic Compilation from High-Level Biologically-Oriented Programming Language to Genetic Regulatory Networks. PLoS ONE 6(8): e22490. doi:10.1371/journal.pone.0022490 Editor: Eshel Ben-Jacob, Tel Aviv University, Israel Received: March 15, 2011; Accepted: June 22, 2011; Published: August 5, 2011 Copyright: © 2011 Beal et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Funding: NIH R01: “Engineering Synthetic Multicellular Systems” (Grant # 7R01GM74712-5) ( This funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Competing interests: The authors have read the journal's policy and have the following conflicts: Jacob Beal is an employee of BBN Technologies. Introduction. Bringing It Together with RNA. Organization of Intracellular Reactions with Rationally Designed RNA Assemblies. Methods in Enzymology : Multiplexed Genome Engineering and Genotyping Methods : : Applications for Synthetic Biology and Metabolic Engineering. Chip chips away at the cost of a genome. Precise Manipulation of Chromosomes in Vivo Enables Genome-Wide Codon Replacement. Microfluidic technologies for synthetic biology. [Int J Mol Sci. 2011] - PubMed result.
GoldenBraid: An Iterative Cloning System for Standardized Assembly of Reusable Genetic Modules. SCRM paper - revision draft 13 - Powered by Google Docs. Organization of Intracellular Reactions with Rationally Designed RNA Assemblies. Synthetic Physiology. A Synthetic Optogenetic Transcription Device Enhances Blood-Glucose Homeostasis in Mice. Scaling Up Digital Circuit Computation with DNA Strand Displacement Cascades. Synthetic Biology. *Scientists discover double meaning in genetic code* by Anonymous "Scientists have discovered a second code hiding within DNA.
This second code contains information that changes how scientists read the instructions contained in DNA and interpret mutations to make sense of health and disease. Genome scientist Dr. John Stamatoyannopoulos. Read the research paper. The work is part of the Encyclopedia of DNA Elements Project, also known as ENCODE. Since the genetic code was deciphered in the 1960s, scientists have assumed that it was used exclusively to write information about proteins.
“For over 40 years we have assumed that DNA changes affecting the genetic code solely impact how proteins are made,” said Stamatoyannopoulos. The genetic code uses a 64-letter alphabet called codons. The discovery of duons has major implications for how scientists and physicians interpret a patient’s genome and will open new doors to the diagnosis and treatment of disease. Amit_2010. Boston University: Applied BioDynamics Laboratory. Engineering microbial biofuel tolerance. Abstract Many compounds being considered as candidates for advanced biofuels are toxic to microorganisms. This introduces an undesirable trade‐off when engineering metabolic pathways for biofuel production because the engineered microbes must balance production against survival. Cellular export systems, such as efflux pumps, provide a direct mechanism for reducing biofuel toxicity. Tunable_signal. Informing Biological Design by Integration of Systems and Synthetic Biology.
In silico feedback for in vivo regulation of a gene expression circuit : Nature Biotechnology. In silico feedback for in vivo regulation of a gene expression circuit : Nature Biotechnology. In silico feedback for in vivo regulation of a gene expression circuit : Nature Biotechnology.