

The Dawn of Virtual Cell Biology: journal club video available. Today I attended a virtual journal club.
Organized by Stephen Larson of OpenWorm (I talked about that very cool 3D worm project in a tip of the week a while back), it also included a few folks from OpenWorm, and two of the authors of the paper: Jonathan Karr and Jayodita Sanghvi. Jonathan’s cat also showed up at one point Stephen did a part in the beginning that was a presentation of the main features of the paper, and the context of the work in biology today.
I think he had quite a similar take on it to what I was describing in my tip this week: modeling types of papers cause a bit of drama and resistance in the biology community; and that this relied on so much previous work in hundreds of publications and lots of curated databases. But he also stressed the importance of this work in moving this field forward. The discussion part was terrific. Anyway, the video is available and you can watch. I really enjoyed the virtual journal club idea and format. Noncanonical Wnt Signaling Maintains Hematopoietic Stem Cells in the Niche. To view the full text, please login as a subscribed user or purchase a subscription.

Click here to view the full text on ScienceDirect. Nuclear Envelope Budding Enables Large Ribonucleoprotein Particle Export during Synaptic Wnt Signaling. Figure 1 Subnuclear Localization of DFz2C and LamC at Larval Muscle Nuclei and Defective NMJs in lamC Mutants (A) LamC and DFz2C labeling (deconvolved) of muscle nucleus containing a DFz2C/LamC focus (box; enlarged in right panels) localized to the nuclear periphery (arrowhead in XZ plane).

Arrows indicate DFz2C granule within the LamC framework-like structure. Structure - Visualizing the Determinants of Viral RNA Recognition by Innate Immune Sensor RIG-I. To view the full text, please login as a subscribed user or purchase a subscription.

Click here to view the full text on ScienceDirect. Figure 1. Snapshot: Endogenous RNAi Pathways. Redirection page. Transcriptional Silencing of Transposons by Piwi and Maelstrom and Its Impact on Chromatin State and Gene Expression. Structural Switch of Lysyl-tRNA Synthetase between Translation and Transcription. To view the full text, please login as a subscribed user or purchase a subscription.
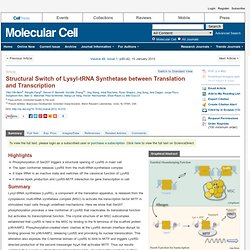
Click here to view the full text on ScienceDirect. Figure 1 Structural Basis for Reserving LysRS in the MSCdirect (A and B) Two orthogonal views of the human LysRS:p38/AIMP2 complex structure. Sequence of p38/AIMP2 is color coded as in (C). (C) Sequence alignment of the N terminus of p38/AIMP2 from Drosophila to human. (D and E) The interface of the LysRS:p38/AIMP2 complex. (F) The functional LysRS:p38/AIMP2 complex. RNase H2-Initiated Ribonucleotide Excision Repair. To view the full text, please login as a subscribed user or purchase a subscription.

Click here to view the full text on ScienceDirect. Figure 1 Ribonucleotide Incorporation during DNA Synthesis (A) Primed RPA-coated SS-M13mp18 DNA was fully replicated by either Pol δ or Pol ε with PCNA under standard conditions, with or without rNTPs ( Experimental Procedures ). Daddy Issues: Paternal Effects on Phenotype. To view the full text, please login as a subscribed user or purchase a subscription.
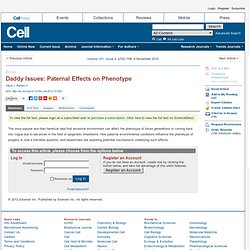
Click here to view the full text on ScienceDirect. Figure 1 Potential Mechanisms Underlying Paternal Environmental Effects on Offspring Phenotype. Capturing the Cloud: UAP56 in Nuage Assembly and Function. Transcriptome-wide Regulation of Pre-mRNA Splicing and mRNA Localization by Muscleblind Proteins. To view the full text, please login as a subscribed user or purchase a subscription.

Click here to view the full text on ScienceDirect. Figure 1. Epigenetic Reprogramming and Small RNA Silencing of Transposable Elements in Pollen. To view the full text, please login as a subscribed user or purchase a subscription.

Click here to view the full text on ScienceDirect. Specialized piRNA Pathways Act in Germline and Somatic Tissues of the Drosophila Ovary. To view the full text, please login as a subscribed user or purchase a subscription.
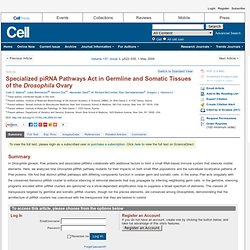
Click here to view the full text on ScienceDirect. Figure 1 Maternal Deposition of piRNAs Defines Somatic and Germline piRNA Pathways. Poxviruses Deploy Genomic Accordions to Adapt Rapidly against Host Antiviral Defenses. To view the full text, please login as a subscribed user or purchase a subscription. Click here to view the full text on ScienceDirect. Figure 1.
The Drosophila HP1 Homolog Rhino Is Required for Transposon Silencing and piRNA Production by Dual-Strand Clusters. To view the full text, please login as a subscribed user or purchase a subscription. Click here to view the full text on ScienceDirect. Figure 1 DNA Damage Signaling in rhi Mutants (A–C) Mutations in mnk, which encodes the DNA damage signaling kinase Chk2, suppress the Gurken and Vasa protein localization defects in rhi mutants. (A) In a stage 9 wild-type oocyte, Grk (blue) is localized at the dorsal anterior cortex near the oocyte nucleus and Vas (red) is localized at the posterior cortex. (B) In rhi egg chambers, this localization pattern is lost, with Grk and Vas dispersed throughout the oocyte. Developmental Cell - The TDRD9-MIWI2 Complex Is Essential for piRNA-Mediated Retrotransposon Silencing in the Mouse Male Germline. Figure 4. Distinct Argonaute-Mediated 22G-RNA Pathways Direct Genome Surveillance in the C. elegans Germline.
To view the full text, please login as a subscribed user or purchase a subscription. Click here to view the full text on ScienceDirect. Figure 1 Hypomorphic Alleles of drh-3 Are RNAi Deficient and Temperature Sensitive (A) Schematic of the drh-3 gene structure. Top panel, conserved DExH and HELICc domains and drh-3 lesions; bottom panel, four missense alleles (indicated) map to the HELICc domain. TOR-tured Yeast Find a New Way to Stand the Heat. Should I Stay or Should I Go? Chromodomain Proteins Seal the Fate of Heterochromatic Transcripts in Fission Yeast. Protection from Feed-Forward Amplification in an Amplified RNAi Mechanism. To view the full text, please login as a subscribed user or purchase a subscription.
Click here to view the full text on ScienceDirect. The Loop Position of shRNAs and Pre-miRNAs Is Critical for the Accuracy of Dicer Processing In Vivo. Clocks, Metabolism, and the Epigenome. PIDD Death-Domain Phosphorylation by ATM Controls Prodeath versus Prosurvival PIDDosome Signaling. Figure 4 ATM Phosphorylates PIDD on T788 during CS Apoptosis. LincRNA-p21 Suppresses Target mRNA Translation. A CXCL1 Paracrine Network Links Cancer Chemoresistance and Metastasis. Figure S2 CXCL1/2 Inhibition in Cancer Cells Does Not Affect Tumor Angiogenesis or Proliferation but Affects Myeloid Recruitment, Related to Figure 2 . Fluorescence-Based Sensors to Monitor Localization and Functions of Linear and K63-Linked Ubiquitin Chains in Cells. Discrete Small RNA-Generating Loci as Master Regulators of Transposon Activity in Drosophila. Figure 1. Structural and Mechanistic Basis for the Inhibition of Escherichia coli RNA Polymerase by T7 Gp2.
<div class="jsDisabled"> To use the Enhanced view of this article, please enable JavaScript on in your browser and refresh the page. </div> Article Search for articles by this author. Promoters Recognized by Forkhead Proteins Exist for Individual 21U-RNAs. Transcript Dynamics of Proinflammatory Genes Revealed by Sequence Analysis of Subcellular RNA Fractions. Redirection page. U1 snRNP Determines mRNA Length and Regulates Isoform Expression. Current Biology - Slicing-Independent RISC Activation Requires the Argonaute PAZ Domain. Direct Regulation of tRNA and 5S rRNA Gene Transcription by Polo-like Kinase 1.
AUF1/HnRNP D RNA Binding Protein Functions in Telomere Maintenance. Histone Variant H2A.Bbd Is Associated with Active Transcription and mRNA Processing in Human Cells. Extrathymic Generation of Regulatory T Cells in Placental Mammals Mitigates Maternal-Fetal Conflict. PiRNAs Can Trigger a Multigenerational Epigenetic Memory in the Germline of C. elegans. Cancer Epigenetics: From Mechanism to Therapy. U1 snRNA Rewrites the “Script” MCM8- and MCM9-Deficient Mice Reveal Gametogenesis Defects and Genome Instability Due to Impaired Homologous Recombination. Tissue-Specific Splicing of Disordered Segments that Embed Binding Motifs Rewires Protein Interaction Networks. Short Structured RNAs with Low GC Content Are Selectively Lost during Extraction from a Small Number of Cells. A Detour to Mature. Tissue-Specific Alternative Splicing Remodels Protein-Protein Interaction Networks. Retraction Notice to: Cell Adhesion-Dependent Control of MicroRNA Decay.
PiRNAs Initiate an Epigenetic Memory of Nonself RNA in the C. elegans Germline. C. elegans piRNAs Mediate the Genome-wide Surveillance of Germline Transcripts. RNA Polymerase Backtracking in Gene Regulation and Genome Instability. A Movie of RNA Polymerase II Transcription. Expressed Pseudogenes in the Transcriptional Landscape of Human Cancers. The Akt-SRPK-SR Axis Constitutes a Major Pathway in Transducing EGF Signaling to Regulate Alternative Splicing in the Nucleus. Intrinsic Nucleic Acid-Binding Activity of Chp1 Chromodomain Is Required for Heterochromatic Gene Silencing. Bidirectional Control of mRNA Translation and Synaptic Plasticity by the Cytoplasmic Polyadenylation Complex.
Dynamic Tyrosine Phosphorylation Modulates Cycling of the HSP90-P50CDC37-AHA1 Chaperone Machine. Structural Basis for Telomerase RNA Recognition and RNP Assembly by the Holoenzyme La Family Protein p65. Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3′ UTRs and near Stop Codons. Insights into RNA Biology from an Atlas of Mammalian mRNA-Binding Proteins. Getting RNA and Protein in Phase. The Structure of Human Argonaute-2 in Complex with miR-20a. The mRNA-Bound Proteome and Its Global Occupancy Profile on Protein-Coding Transcripts. Rrp6p Controls mRNA Poly(A) Tail Length and Its Decoration with Poly(A) Binding Proteins. HP1Swi6 Mediates the Recognition and Destruction of Heterochromatic RNA Transcripts.
Current Biology - Birth of New Spliceosomal Introns in Fungi by Multiplication of Introner-like Elements. Insights into RNA Biology from an Atlas of Mammalian mRNA-Binding Proteins. Nascent-Seq Indicates Widespread Cotranscriptional RNA Editing in Drosophila. MicroRNA Factory: RISC Assembly from Precursor MicroRNAs. mRNA Decay Factor AUF1 Maintains Normal Aging, Telomere Maintenance, and Suppression of Senescence by Activation of Telomerase Transcription. Role of the SEL1L:LC3-I Complex as an ERAD Tuning Receptor in the Mammalian ER. Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3′ UTRs and near Stop Codons. Cytoplasmic Assembly and Selective Nuclear Import of Arabidopsis ARGONAUTE4/siRNA Complexes. The Sam68 STAR RNA-Binding Protein Regulates mTOR Alternative Splicing during Adipogenesis. The Poly(A)-Binding Protein Nuclear 1 Suppresses Alternative Cleavage and Polyadenylation Sites. Inactivation of Conserved C. elegans Genes Engages Pathogen- and Xenobiotic-Associated Defenses.
Staufen1-Mediated mRNA Decay Functions in Adipogenesis. Precursor MicroRNA-Programmed Silencing Complex Assembly Pathways in Mammals. Nuclear Envelope Budding Enables Large Ribonucleoprotein Particle Export during Synaptic Wnt Signaling. Hsp90 Globally Targets Paused RNA Polymerase to Regulate Gene Expression in Response to Environmental Stimuli. Cell-free Formation of RNA Granules: Bound RNAs Identify Features and Components of Cellular Assemblies.
RNP Export by Nuclear Envelope Budding. Species-Dependent Posttranscriptional Regulation of NOS1 by FMRP in the Developing Cerebral Cortex. Cell-free Formation of RNA Granules: Low Complexity Sequence Domains Form Dynamic Fibers within Hydrogels. Decapping Goes Nuclear. Translational Homeostasis via the mRNA Cap-Binding Protein, eIF4E. mRNA Decapping Factors and the Exonuclease Xrn2 Function in Widespread Premature Termination of RNA Polymerase II Transcription. A Role for tRNA Modifications in Genome Structure and Codon Usage. A Role for Neuronal piRNAs in the Epigenetic Control of Memory-Related Synaptic Plasticity. DICER1 Loss and Alu RNA Induce Age-Related Macular Degeneration via the NLRP3 Inflammasome and MyD88. A Long ncRNA Links Copy Number Variation to a Polycomb/Trithorax Epigenetic Switch in FSHD Muscular Dystrophy.
R Loops: From Transcription Byproducts to Threats to Genome Stability. Reduced Expression of Ribosomal Proteins Relieves MicroRNA-Mediated Repression. CRISPR Immunity Relies on the Consecutive Binding and Degradation of Negatively Supercoiled Invader DNA by Cascade and Cas3. Reconstitution of an Argonaute-Dependent Small RNA Biogenesis Pathway Reveals a Handover Mechanism Involving the RNA Exosome and the Exonuclease QIP. Small RNAs as Guardians of the Genome. Developmental Cell - MITOPLD Is a Mitochondrial Protein Essential for Nuage Formation and piRNA Biogenesis in the Mouse Germline.
The CRISPR System: Small RNA-Guided Defense in Bacteria and Archaea. 3′ End Formation of PIWI-Interacting RNAs In Vitro. Collapse of Germline piRNAs in the Absence of Argonaute3 Reveals Somatic piRNAs in Flies. RNA-Guided RNA Cleavage by a CRISPR RNA-Cas Protein Complex.