

Labeling Oligonucleotides and Nucleic Acids—Section 8.2. To facilitate the preparation of optimally labeled nucleic acids, we offer several modified nucleotides, as well as a wide selection of nucleic acid labeling kits that contain reactive versions of Molecular Probes fluorophores.
Our available technologies include: ChromaTide dUTP, ChromaTide OBEA-dCTP and ChromaTide UTP nucleotides, which provide researchers with a large selection of fluorophore- and hapten-labeled nucleotides that can be enzymatically incorporated into DNA or RNA probes for FISH (fluorescence in situ hybridization), DNA arrays/microarrays and other hybridization techniques. MIQE. MIQE: Minimum Information for Publication of Quantitative Real-Time PCR Experiments The RDML file format is a recommended element in the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines (Bustin et al., Clinical Chemistry, 2009).

The aim of MIQE, coordinated by a group of research-active scientists and coordinated under the umbrella of MIBBI (Minimum Information for Biological and Biomedical Investigations) is to provide authors, reviewers and editors specifications for the minimum information that must be reported for a qPCR experiment in order to ensure its relevance, accuracy, correct interpretation and repeatability. A checklist, which should be submitted along with the paper, is available for authors in preparing a manuscript employing qPCR Download the MIQE paper Download the MIQE checklist Letter to Editors inviting them to adopt the MIQE standard for their papers Checklist for Quantitative PCR Assays. Large intergenic non-coding RNAs (lincRNAs) are emerging as key regulators of diverse cellular processes.
Determining the function of individual lincRNAs remains a challenge. Recent advances in RNA sequencing (RNA-Seq) and computational methods allow for an unprecedented analysis of such transcripts. In this work, we present an integrative approach to define a reference catalogue of over 8,000 human lincRNAs. Our catalogue unifies previously existing annotation sources with transcripts we assembled from RNA-Seq data collected from ~4 billion RNA-Seq reads across 24 tissues and cell types. We characterize each lincRNA by a panorama of more than 30 properties, including sequence, structural, transcriptional, and orthology features. This site includes the catalog of the Human Body Map lincRNAs and TUCP transcripts as well as all RNA-Seq alignment files and raw de-novo transcriptome assemblies.
If you have any questions please address them to Moran nmcabili at fas.harvard.edu.
SNPs. Patent WO2002045472A2 - SEQUENCE SPECIFIC AND SEQUENCE NON-SPECIFIC METHODS AND MATERIALS FOR cDNA ... - Google Patenter. SEQUENCE SPECIFIC AND SEQUENCE NON-SPECIFIC METHODS AND MATERIALS FOR cDNA NORMALIZATION AND SUBTRACTION Each of the applications and patents cited in this text, as well as each document or reference cited in each of these applications and patents (including during the prosecution of each issued patent; "application cited documents"), and each of the PCT and foreign applications or patents corresponding to and/or claiming priority from any of these applications and patents, and each of the documents cited or referenced in each of the application cited documents, are hereby expressly incorporated herein by reference in their entirety.

FIELD OF THE INVENTION The present invention discloses methods and materials that efficiently normalize cDNA libraries. The present invention also discloses methods and materials for aiding subtractive/differential hybridization and other normalization procedures. The methods and materials can be packaged in the form of a kit. Soares et al. (1994) Proc. enhancer.lbl.gov. Repeat associated Transcription Start Sites - ZENBU documentation wiki. From ZENBU documentation wiki Case study focused on extracting repetitive elements sub-cellular compartment specific expression from ENCODE K562 cell line analyzed by CAGE.
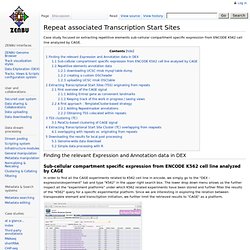
Finding the relevant Expression and Annotation data in DEX Sub-cellular compartment specific expression from ENCODE K562 cell line analyzed by CAGE In order to find all the CAGE experiments related to K562 cell line in encode, we simply go to the "DEX - expression/experiment" tab and type "K562" in the upper right search box. The lower drop down menu allows us the further inspect all the "experiment platforms" under which K562 related experiments have been stored and further filter the results of the "K562" query for a specific experimental platform. Glyph style "express" (simple wiggle-plot like histogram binning) region : "5'end" (only the very first 5'ending base will be graphed) binning : "sum" which will sum up overlapping 5'ends from tags origitating from all the samples selected.
RNA-Seq: A brief history - Seven Bridges Genomics.
RNA. Cloning.