

Microbiome: Expanding the Gut Gene Catalog. MetaHIT project workflow to create a human gut gene catalog from three continents. 1 The microbes that live in our intestines are intimately tied to our health and represent a rich source of unexplored metabolic information.
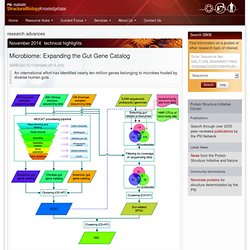
A typical gut community may possess an order of magnitude more genes than are encoded in the genome of its human host. New work by Wang, Bork and colleagues at the Metagenomics of the Human Intestinal Tract (MetaHIT) consortium greatly expands the list of known gut microbial genes in a single high-quality gene catalog, making it an excellent resource for gene function and structural studies.
With the goal of increasing the number and diversity of sampled populations, the MetaHIT consortium sequenced new Danish and Spanish samples and analyzed sequence data from their previous European samples, American samples from the Human Microbiome Project and Chinese samples from a diabetes study. Tal Nawy. Coaxing Rare Codons. Protein-Nucleic Acid Interaction: A Modified SAM to Modify tRNA : PSI-Nature Structural Biology Knowledgebase.
Stick representation of the computationally predicted pose of prephenate (grey) in the CmoA catalytic site.
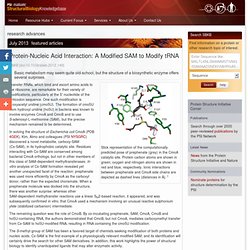
Protein carbon atoms are shown in green; oxygen and nitrogen atoms are shown in red and blue, respectively. Ionic interactions between prephenate and CmoA side chains are depicted as dashed lines (distances in Å). 1 Transfer RNAs, which bind and escort amino acids to the ribosome, are remarkable for their variety of modifications, particularly at the 5′ nucleotide of the anticodon sequence. Viral RNA: a perfect fit for IFIT : PSI-Nature Structural Biology Knowledgebase. Protein Interaction Networks: Morph to Assemble : PSI-Nature Structural Biology Knowledgebase. Family of models that represent the NMR solution structure of the S-state of E. coli IscU.
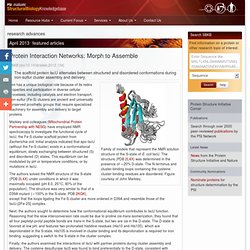
The structure (PDB 2L4X) was determined in the presence of ∼20% D-state. The N-terminus and cluster-binding loops containing the cysteine cluster-binding residues are disordered. Figure courtesy of John Markley. Iron has a unique biological role because of its redox properties and participation in diverse cellular processes, including catalysis and electron transport. Iron-sulfur (Fe-S) clusters are ancient and universally conserved prosthetic groups that require specialized machinery for assembly and delivery to target proteins.
Markley and colleagues (Mitochondrial Protein Partnership with NESG) have employed NMR spectroscopy to investigate the functional cycle of IscU, the Fe-S cluster scaffold protein from Escherichia coli. The authors solved the NMR structure of the S-state (PDB 2L4X) under conditions in which it was maximally occupied (pH 8.0, 25°C, 80% of the population). Michael A. Protein Interaction Networks: Reading Between the Lines : PSI-Nature Structural Biology Knowledgebase. LC-IMS-MS analysis results in signals separated by m/z (y axis) and drift time (x axis).
Doublets are signals from heavy/light peptide pairs, but the triplet (shown above) is from a cross-linked homodimeric peptide (signals are from light-light, light-heavy, heavy-light and heavy-heavy pairs, giving a 1:2:1 ratio). Figure courtesy of Joshua Adkins. Structural proteomics efforts have greatly benefited from the combination of cross-linking and mass spectrometry (MS). In the presence of a cross-linker, adjacent portions of proteins are linked covalently, and subsequent MS analysis identifies those neighboring fragments, providing clues about overall structure.
Complexes with multiple copies of the same protein, however, present the problem of distinguishing intra- from intermolecular cross-links: unless the identified peptides have overlapping, non-repeated sequences, the distinction cannot be made. Irene Kaganman. Crystal Structure and RNA Binding Properties of the RNA Recognition Motif (RRM) and AlkB Domains in Human AlkB Homolog 8 (ABH8), an Enzyme Catalyzing tRNA Hypermodification. Nanopores quantify miRNAs : PSI-Nature Structural Biology Knowledgebase.
Follow the RNA leader : PSI-Nature Structural Biology Knowledgebase. Residues near the gag start codon (AUG, green) form a hairpin in the monomeric RNA, and residues that promote dimerization (DIS, red) are sequestered by base-pairing with U5.
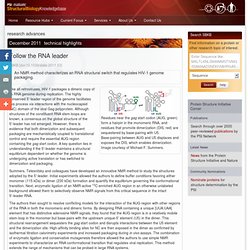
Base-pairing between AUG and U5 displaces and exposes the DIS, which enables dimerization. Image courtesy of Michael F. Summers. Like all retroviruses, HIV-1 packages a dimeric copy of its RNA genome during replication. The highly conserved 5′-leader region of the genome facilitates this process via interactions with the nucleocapsid (NC) domain of the viral Gag polyprotein. Summers, Telesnitsky and colleagues have developed an innovative NMR method to study the structures adopted by the 5′-leader. NMR Detection of Structures in the HIV-1 5′-Leader RNA That Regulate Genome Packaging. An effective and cooperative dimer : PSI-Nature Structural Biology Knowledgebase.
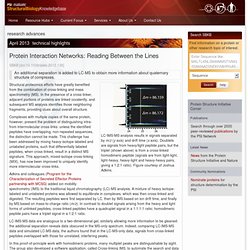
Working model useful in guiding studies of the cooperative binding of NS1A to dsRNA.
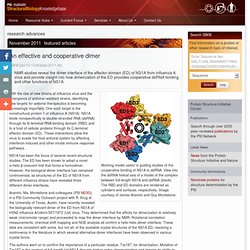
View into the dsRNA helical axis of a model of the complex between full-length NS1A and dsRNA (black). The RBD and ED domains are rendered as cylinders and surfaces, respectively. Image courtesy of James Aramini and Guy Montelione. Structure from sequence : PSI-Nature Structural Biology Knowledgebase. RMDetect uses sequence information to predict the presence of RNA structural modules.
Reprinted from Nature Methods. 1 It is no secret that tertiary structures in RNA are almost, if not equally, as important as the RNA sequence itself. These structures are formed by short- and long-range interactions between the bases in the RNA and often take the form of specific motifs, also referred to as modules. These modules include G-bulges, kink-turns, C-loops and tandem GA/AG loops (tandem GAs). However, although there are tools to predict RNA secondary structure motifs, they are limited by their ineffectiveness to treat the whole range of sequence variations. Now, Cruz and Westhof have generated a new computational tool for detecting RNA structural modules on the basis of the RNA sequence alone.
To validate the utility of RMDetect, the authors tested it against single target sequences as well as multiple sequence alignments. Steve Mason. Home : PSI-Nature Structural Biology Knowledgebase.