

Integrated DNA Technologies. Removal of Shelterin Reveals the Telomere End-Protection Problem. Limitations of silencing at native yeast telomeres : Article : The EMBO Journal. Limitations of silencing at native yeast telomeres : Abstract : The EMBO Journal. Additional Modules for Versatile and Economical PCR-based Gene Deletion and Modification in Saccharomyces cerevisiae. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1.
+ Author Affiliations Abstract The maintenance of transcriptional silencing at HM mating-type loci and telomeres in yeast requires the SIR2, SIR3, and SIR4 proteins, none of which appear to be DNA-binding proteins.

Here we show that SIR3 and SIR4 interact with a carboxy-terminal domain of the silencer, telomere, and UAS-binding protein RAP1. We identified SIR3 and SIR4 in a two-hybrid screen for RAP1-interacting factors and showed that SIR3 interacts both with itself and with SIR4. The interaction between RAP1 and SIR3 can be observed in vitro in the absence of other yeast proteins. Footnotes Copyright © Cold Spring Harbor Laboratory Press.
Distinct Differences in Chromatin Structure at Subtelomeric X and Y' Elements in Budding Yeast. In Saccharomyces cerevisiae, all ends of telomeric DNA contain telomeric repeats of (TG1–3), but the number and position of subtelomeric X and Y' repeat elements vary.

Using chromatin immunoprecipitation and genome-wide analyses, we here demonstrate that the subtelomeric X and Y' elements have distinct structural and functional properties. Y' elements are transcriptionally active and highly enriched in nucleosomes, whereas X elements are repressed and devoid of nucleosomes. In contrast to X elements, the Y' elements also lack the classical hallmarks of heterochromatin, such as high Sir3 and Rap1 occupancy as well as low levels of histone H4 lysine 16 acetylation.
Our analyses suggest that the presence of X and Y' elements govern chromatin structure and transcription activity at individual chromosome ends. Figures. ChIC and ChEC: Genomic Mapping of Chromatin Proteins. To view the full text, please login as a subscribed user or purchase a subscription.
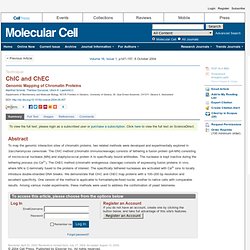
Click here to view the full text on ScienceDirect. Figure 1 Outline of ChIC (A) The major steps of the ChIC procedure are illustrated. (Step 1) Binding of the primary antibody (AB) and subsequent centrifugation-washing steps. (B) Immunoblots of ChIC experiments with fixed crude nuclei derived from cells expressing either the antigen Gbd-myc13 (strain KIY64 transformed with pGBC11-myc13), the antigen Sir3-myc13 (strain KIY67), or no antigen (no myc, strain KIY64). Figure 2. TERRA Promotes Telomere Shortening through Exonuclease 1–Mediated Resection of Chromosome Ends. Author SummaryTelomeres protect chromosome ends from end fusion and end degradation, and they regulate cellular lifespan. Telomerase, a reverse transcriptase, maintains telomere length. The end replication problem and the processing of DNA ends by nucleases cause telomere shortening. Telomeres are transcribed into a long noncoding RNA known as TERRA. ICF syndrome derived patient cells have short telomeres and enriched TERRA.
TERRA inhibits telomerase activity in vitro. Verena Pfeiffer , Joachim Lingner * Swiss Institute for Experimental Cancer Research (ISREC), School of Life Sciences, Frontiers in Genetics National Center of Competence in Research, Ecole Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland Abstract Top The long noncoding telomeric repeat containing RNA (TERRA) is expressed at chromosome ends.
Mutually Exclusive Binding of Telomerase RNA and DNA by Ku Alters Telomerase Recruitment Model. To view the full text, please login as a subscribed user or purchase a subscription.

Click here to view the full text on ScienceDirect. Figure 1 Truncations within the Ring of Ku80 Impair NHEJ and Gene Silencing via Telomere Position Effect (A) Internal deletions within the loop that slides over the dsDNA were designed on the basis of the crystal structure of human Ku ( Walker et al., 2001 ). (B) Using a strain with an engineered HO endonuclease cut-site, the Ku mutants, along with the WT strain and Δku80, were streaked onto plates containing either glucose or galactose. (C) Western blot of the mutant yeast protein content after 20 generations. (D) Silencing assay for the URA3 gene at 20 generations. See also Figure S1 . Figure 2 Mutations within Ku's DNA-Binding Loop Reduce Telomere Length. Cohabitation of insulators and silencing elements in yeast subtelomeric regions : Abstract : The EMBO Journal. Introduction Eukaryotic genomes are thought to be organized into chromatin domains in which a number of processes such as transcription, replication and splicing are regulated autonomously and locally (reviewed in Lamond and Earnshaw, 1998).
Chromosomal domains may simply arise as a consequence of their autonomous function or may be delimited actively by specialized boundary elements. While abundant evidence supports the existence of such elements in Drosophila and mammals (reviewed in Gerasimova and Corces, 1996), boundary elements have not been described as yet in yeast. A most conspicuous feature of the yeast genome compared with that of multicellular organisms is its compactness (Dujon, 1996). Non‐coding DNA occupies very little space and genes lie in apparent promiscuity. Key components of silenced chromatin in S.cerevisiae are the silent information regulators Sir2p, Sir3p and Sir4p, as well as histones H3 and H4 (for reviews, see Sherman and Pillus, 1997; Lustig, 1998). Results. FEBS Letters - When the caps fall off: Responses to telomere uncapping in yeast.