

Cereal Chemistry Journal - 82(6):706 - Abstract. A Rapid CTAB DNA Isolation Technique Useful for RAPD Fingerprinting and Other PCR Applications. A Rapid CTAB DNA Isolation Technique Useful for RAPD Fingerprinting and Other PCR Applications (BioTechniques 1993 article Vol. 14(5):748-749) C.

Neal Stewart, Jr., and Laura E. Via Biology Department Virginia Polytechnic Institute and State University Blacksburg, VA 24061 Internet:nstewart@uga.cc.uga.edu Many DNA isolation techniques widely employed by plant molecular biologists use a CTAB (hexadecyltrimethylammonium bromide) extraction buffer coupled with reusable tissue homogenization systems such as a mortar and pestle (3,4,10). The procedure presented is a modification of the Doyle and Doyle CTAB method (4) scaled to fit in microcentrifuge tubes with reagents (6) added to increase separation of polysaccharides from the DNA. For the CTAB mini prep method, an Eppendorf 1000 l plastic pipette tip that had been pushed onto a deburring tool mounted on a cordless drill served as the homogenizing pestle.
DNA from both procedures was quantified using a minifluorometer Hoefer TKO 100). Beta-kafirin [Sorghum bicolor] - Protein - NCBI. Gamma-kafirin [Sorghum bicolor] - Protein - NCBI. Delta kafirin [Sorghum bicolor] - Protein - NCBI. Alpha Kafirin.
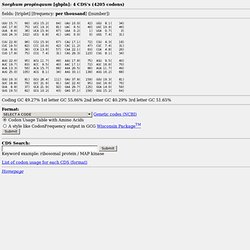
Codon usage table. Sorghum propinquum [gbpln]: 4 CDS's (4205 codons) fields: [triplet] [frequency: per thousand] ([number]) Coding GC 49.27% 1st letter GC 55.86% 2nd letter GC 40.29% 3rd letter GC 51.65% List of codon usage for each CDS (format) Homepage.
Codon usage. An expanded codon table showing the relative frequence that different codons are used in E. coli genes is shown below.

REFERENCE: Modified from Maloy, S., V. Stewart, and R. Taylor. 1996. Genetic analysis of pathogenic bacteria. Cold Spring Harbor Laboratory Press, NY. Click here to return to the Microbial Genetics Home Page. Codon Plus E.Coli. SorghumCodonUsage.xlsx. Expression Pattern of the Alpha-Kafirin Promoter Coupled with a Signal Peptide from Sorghum bicolor L. Moench.
Godwin_molecular_characterization. Temporal and spatial expression analysis of gam... [Mol Biol Rep. 2008. Gamma Kafirin Sequence. Structural characterization and promoter activ... [Mol Gen Genet. 1994. Kafirin Structure and Functionality. Sorghum Kafirin Info. Mutant Kafirin Digestible. Agrobacterium-mediated sorghum transformation. [Plant Mol Biol. 2000. Analysis of kafirin promoter activity in tran... [Plant Mol Biol. 1996. Agrobacterium Sorghum Transformation. Agrobacterium-mediated sorghum transformation using simple binary vectors (Lu et al. 2009.

PCTO: J Plant Biotechnol 99:97–108) Note: we have made new improvements in sorghum regeneration and transformation and we are now using sorghum genotype TX430 for all transformation services. Our new transformation process improved from the protocol below will be publicized at a later time. Initiation of Agrobacterium tumefaciens culture: Streak out Agrobacterium culture from -80°C glycerol stock onto YEP medium plate containing appropriate antibiotics and place the plate at 25°C for 4 days. Embryo isolation, infection: Sterilize kernels from greenhouse-grown sorghum variety P898012 with 50% bleach and a few drops (0.1%) Tween-20 for 30 min with vacuum using large desiccators for the vacuum. Tannin. Tannin powder (mixture of compounds).
Tannins have molecular weights ranging from 500 to over 3,000[3] (gallic acid esters) and up to 20,000 (proanthocyanidins). Structure and classes of tannins[edit] There are three major classes of tannins:[4] Shown below are the base unit or monomer of the tannin. Particularly in the flavone-derived tannins, the base shown must be (additionally) heavily hydroxylated and polymerized in order to give the high molecular weight polyphenol motif that characterizes tannins. Typically, tannin molecules require at least 12 hydroxyl groups and at least five phenyl groups to function as protein binders.
Pseudo tannins[edit] Sorghum kafirin interaction with various phenolic compounds - Emmambux - 2003 - Journal of the Science of Food and Agriculture.